Today, we’ll be using OHDSI tools to develop and evaluate cohort definitions for the phenotype target of Type 2 diabetes mellitus (T2DM).
Clinical description:
The American Diabetes Association (ADA) “Standards of Medical Care in Diabetes” is a tremendous resource to learn more about diabetes for those interested. It classifies diabetes into "the following general categories:
- Type 1 diabetes (due to autoimmuneb-cell destruction, usually leading to absolute insulin deficiency, including latent autoimmune diabetes of adulthood)
- Type 2 diabetes (due to a progressive loss of adequate b-cell insulin secretion frequently on the background of insulin resistance)
- Specific types of diabetes due to other causes, e.g.,monogenic diabetes syndromes (such as neonatal diabetes and maturity-onset diabetes of the young), diseases of the exocrine pancreas (such as cystic fibrosis and pancreatitis), and drug- or chemical-induced diabetes (such as with glucocorticoid use, in the treatment of HIV/AIDS, or after organ transplantation)
- Gestational diabetes mellitus (diabetes diagnosed in the second or third trimester of pregnancy that was not clearly overt diabetes prior to gestation)"
It provides objective diagnostic criteria based on readily-accessible laboratory measures used in routine practice. “Diabetes can be diagnosed based on plasma glucose criteria, either the fasting plasma glucose (FPG) value or the 2-h plasma glucose (2-h PG) value during a 75-g oral glucose tolerance test (OGTT), or A1C criteria”
The epidemiology and disease natural history of T2DM has been extensively characterized in the literature. Common symptoms of T2DM onset include thirst, frequent urination, weight loss. Common ‘risk factors’ include age, obesity, hypertension and hyperlipidemia. Management of T2DM can include lifestyle modifications, including diet and exercise, as well as pharmacologic treatment (with notable drugs including metformin, Sulfonylureas, Sodium Glucose Co-Transporter 2 (SGLT2) inhibitors, Glucagon-like Peptide-1 Receptor Agonists (GLP1RA), Dipeptidyl peptidase-4 inhibitor (DPP4i), Thiazolidinediones, and insulin). Long-term complications associated ofT2DM can include cardiovascular events (ischemic heart disease, stroke), diabetic retinopathy, kidney failure, and amputation. The incidence of T2DM has been increasing over time. Current prevalence estimates of ~6% in the general population, but vary considerably across countries around the world.
Cohort definitions to evaluate
In this exercise, I created 3 definitions to review:
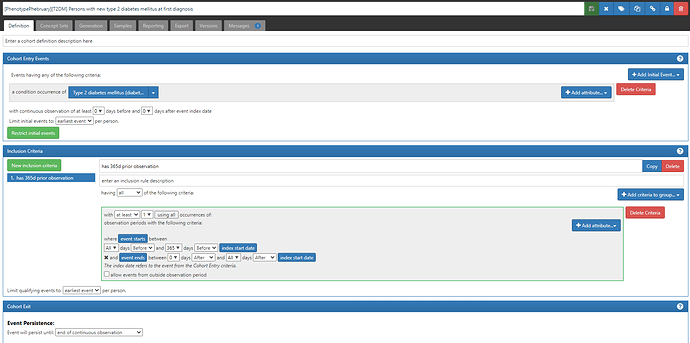
- “Persons with new type 2 diabetes mellitus at first diagnosis” (ATLAS-phenotype link here).
This is a ‘simple’ approach to a phenotype definition, as we commonly see in many observational database studies of T2DM. Namely, we define a person’s status based only on a diagnosis code, as found in the CONDITION_OCCURRENCE table. To find ‘new’ cases of T2DM, we apply two logical devices: 1) we limit the cohort entry events to ‘earliest event’ per person, and 2) we add an inclusion criteria requiring at least 365 days of prior observation (by requiring that the entry event occurs within an observation period whose observation period start is more than 365d before the entry event start and end is sometime afterwards). {Side note, we could have applied the observation period restriction within the entry event criteria, but we would have needed to also specify that the condition occurrence was ‘first time in history’. Implementing it as shown here has the added benefit that the attrition table shows you how many patients were ‘lost’ by imposing the 365d prior observation requirement, which effectively tells you how many ‘prevalent’ T2DM patients the database had who wouldn’t qualify as ‘incident’ cases}
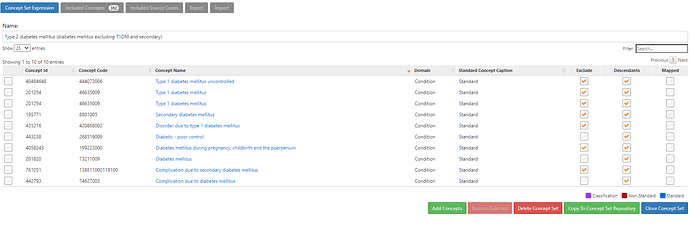
Here, we have to define a conceptset expression to represent the clinical idea of ‘Type 2 diabetes mellitus’. And specifically, as it shows in the name, we’ve done this by identifying ‘diabetes mellitus excluding T1DM and secondary’. An insight here is that there are many standard concepts and source codes that do not distinguish the specific classification of ‘diabetes mellitus’, so we need to decide whether to include or exclude them in our T2DM definition. Notable examples of non-specific standard concepts that are highly prevalent in our OHDSI network include ‘Diabetes mellitus’, ‘Complication due to diabetes mellitus’ and ‘Diabetic-poor control’. Note also that you can’t solely rely on the SNOMED hierarchy to drag in all diabetes-related concepts. Indeed, ‘Diabetes mellitus’, ‘Complications due to diabetes mellitus’ and ‘Diabetic-poor control’ are not part of the same ancestry, in that none are descendants of each other. This conceptset uses these three concepts, plus their descendants, and then excludes concepts underneath of them which we don’t want, like ‘Type 1 diabetes mellitus’, ‘Disorder due to Type 1 diabetes mellitus’ and ‘Secondary diabetes mellitus’ (note, this roughly conforms to the ADA classification above). Finally, I’ll highlight that this succinct conceptset expression, with its 10 concepts, 3 with descendants, and 7 with excluded descendants, that conceptset resolves into an included concept list of 362 standard concepts, that vast majority of which have been observable in at least one database within the OHDSI network. And that 362 standard concepts have 2,177 mapped source codes, including 70 ICD9CM codes, 42 ICD10 codes, 481 ICD10CM codes, 271 Read codes. Imagine having to find all 362 concepts and 2,177 source codes yourself through some primitive string search…good luck! Thank you OHDSI vocabulary team and the ATLAS development team for making our lives so much easier!
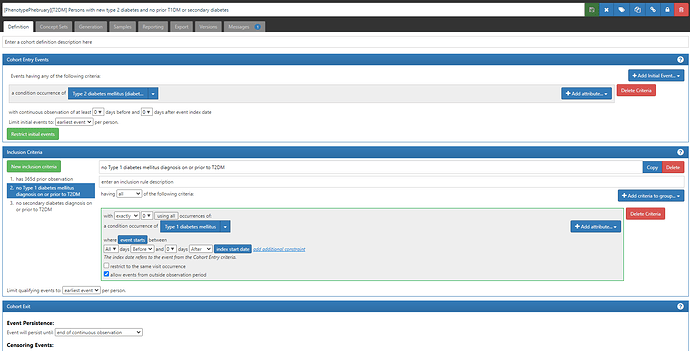
- “Persons with new type 2 diabetes and no prior T1DM or secondary diagnosis” (ATLAS-phenotype link here)
Note, the difference between #1 and #2 is the addition of two new inclusion criteria: ‘no Type 1 diabetes mellitus on or prior to T2DM’ and ‘no secondary diabetes diagnosis on or prior to T2DM’. Each of these inclusion criteria required their own conceptsets to represent T1DM and secondary diabetes, respectively. The logic for using those concepts is to require that exactly 0 condition occurrence records are observed all time prior through the index date, including for events that fall outside the observation period. Even though it has more criteria, this definition relies only on the CONDITION_OCCURRENCE table to identify patients and determine cohort start dates.
This definition is most akin to logic we have currently proposed to use with the LEGEND-T2DM study (protocol is here). The difference there is that we have modeled new user drug cohorts, and the requirement of the indication is represented as inclusion criteria.
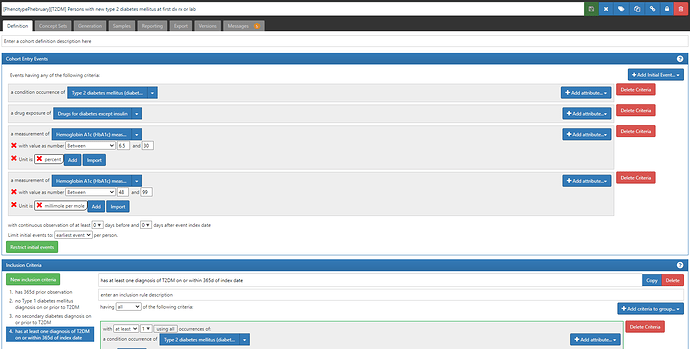
- “Persons with new type 2 diabetes mellitus at first dx rx or lab” (ATLAS-phenotype link here)
This definition uses conditions, drugs, and measurement values to identify the ‘earliest’ entry event, and then adds an inclusion criteria that all persons must have a diagnosis of T2DM on or within the 365d after the index date.
First, note the cohort entry events had 4 components: 1) condition occurrence of ‘Type 2 diabetes mellitus’ (same as in other two definitions), 2) drug exposure of ‘drugs for diabetes except insulin’, 3) measurement of ‘Hemoglobin A1C’ with a value > 6.5 and unit of ‘percent’,or 4) measurement of ‘Hemoglobin A1C’ with a value > 48 and unit of ‘millimole per mole’.
A couple ‘lessons learned the hard way’ to point out here. 1) I specified the value as a range (e.g. for %, it’s ‘between 6.5 and 30’, and this is because we had seen some database have data quality issues with measurement values that could get swept in if not bounded (ex: if a database had values with omitted decimal points, then 60 instead of 6.0 would be greater than 6.5, but not what we want). 2) you have to specify units for measurements, unless you know that all databases are uniform or the measurement can’t have alternative units or is truly unitless. This will have the consequence that some databases that have values but no units will not be able to apply this criteria, but the alternative is far worse: assuming the values follow some distribution when you could be on the completely wrong scale (in this case, confusing % with mmol/mol). 3) if you want to still require the diagnosis code, you can include it in the entry event AND in an inclusion criteria. This way, a person can qualify if EITHER their first entry event is a diagnosis OR their first entry event is a drug or measurement but then later they have a diagnosis. This trick comes in handy when we are trying to correct for index date misspecification that can arise when we only use diagnosis codes (more on that later).
So, without looking at data, we can think about the logic of these three definitions to understand how they should play out. #1 is the superset, with #2 being a subset of #1 that meets 2 additional criteria, and #3 being the same people as #2, except that some persons may be excluded because the index date correction may eliminate some prevalent cases that were captured as ‘incident’ in #2. Note, definition #3, while it is using drugs and measurements also, because it requires a diagnosis, it shouldn’t be a broader definition. (A reasonable alternative one could consider to evaluate is what if one didn’t impose a requirement of a diagnosis, relying ONLY on drug exposures OR measurement values).
Phenotype evaluation using CohortDiagnostics
We used CohortDiagnostics to evaluate these cohorts. The results are available for your review at: https://data.ohdsi.org/phenotypePhebruary/
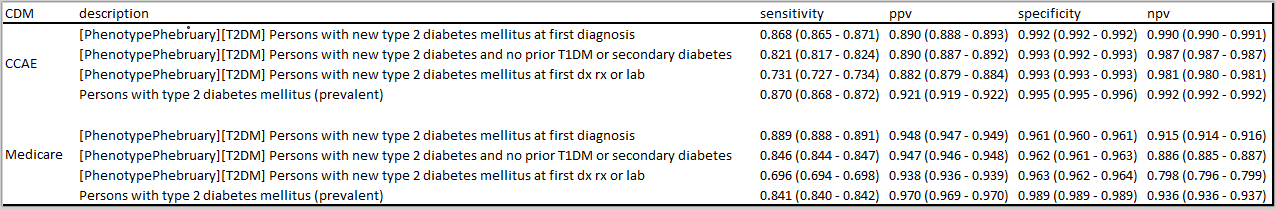
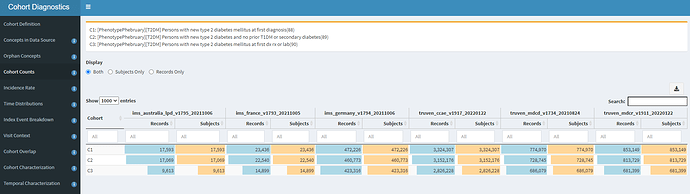
The three cohort definitions described above were applied to six databases in this initial evaluation: 3 US administrative claims datasets : IBM MarketScan Commercial Claims and Encounters (CCAE), IBM MarketScan Multi-state Medicaid (MDCD), and IBM MarketScan Medicare Supplemental Beneficiaries (MDCR); the Iqvia Disease Analyzer - Germany; Iqvia Disease Analyzer - France; and Iqvia LPD - Australia. The cohort counts for all definitions against all databases is shown below.
One of the first observations that you can note from this: Imposing the additional inclusion criteria of ‘no T1DM or secondary diabetes’ in definition C2 had a very small impact on the cohort count, as compared to the original C1 definition. For example, in CCAE, we dropped from 3.32m patients to 3.15m patients (~5% loss). This is directionally aligned with the ADA statement that ~90% of all diabetes cases are T2DM. What was a bit more surprising to me was that we see an additional ~10% drop from C2 to C3 (the definition where we allow entry events based on diagnoses, drugs or measurements). This suggests that many ‘qualifying incident’ patients in C2 may have actually been prevalent T2DM patients, insofaras they had prior diabetes drug use OR elevated HbA1c before their diagnosis (and within the 365d observation period). This is our first clue that there may be some index date misspecification when using T2DM diagnoses to find new cases.
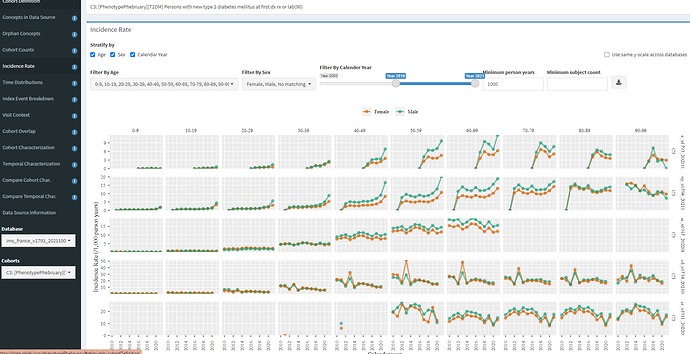
Using the Incidence Rate tab, we can see interesting patterns by age, sex, and index year.
In line with the ADA description, we can see that T2DM incidence increases with age with particular growth from 30 to 40 to 50. We also see, across all 5 databases shown here, the incidence is higher in men (as it expected, both on its own, and due to association with other risk factors, like obesity and hypertension). When we look over index year, we see some interesting patterns. The most notable one for me is MDCD, where there is a clear pronounced spike in incidence in 2013, across all age/sex strata, and this spike is not observed in any other database. While I do not know for sure, I suspect this has to do with states rolling out the Medicaid Incentives for Prevention of Chronic Diseases (MIPCD) in 2013.
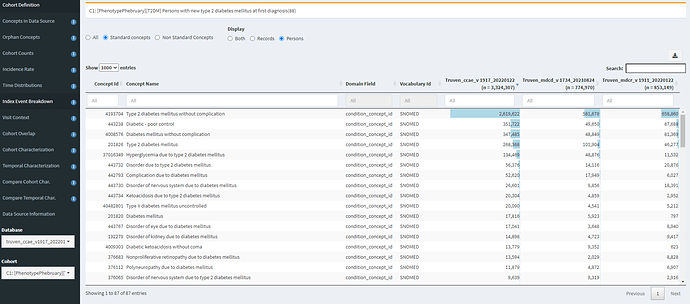
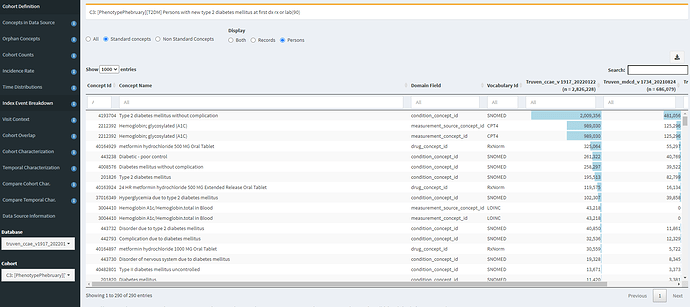
The Index Event Breakdown tab allows you to understand which concept was observed on the index date (that is, what concept was truly the ‘entry event’). I find it useful to look at the C1 definition (based on diagnosis only):
Here, we can see that the main qualifying concept in CCAE and MDCD is ‘Type 2 diabetes mellitus without complication’. However, its interesting to see that other concepts in the top 10 include ‘diabetic-poor control’ (that concept that we might miss if only using the SNOMED hierarchy or a string search for diabetes), ‘Complication due to diabetes mellitus’ (non-specific to T2DM) and various specfic complications that one might have imagined would follow the initial onset of T2DM (such as ‘disorder of eye’, ‘disorder of kidney’, ‘polyneuropathy’).
Now, juxtapose that against C3, where we index on earliest of diagnosis, drug or measurement:
Here, we see that HbA1c and metformin exposure are near the top of the list of entry events. And specific complications are now further down the list and less prevalent (but still do occur as the first observed event).
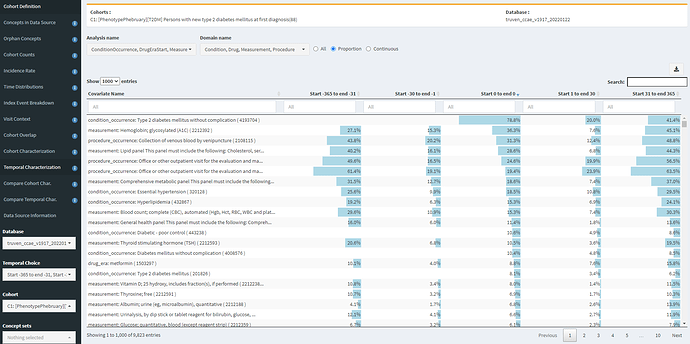
The Temporal Characterization tab is quite useful for identifying specificity errors and index date misspecification errors. First, I show here looking at CCAE for definition C1 (diagnosis only), with the table sorted by feature prevalence on day 0.
We see that a T2DM diagnosis is the most common element (as to be expected), and that 36.3% of people have a HbA1c measurement on day 0, which is likely in line with general expectations. However, we see that this cohort also had 15% having HbA1c measurement in prior 30d and 27% having HbA1c in the 11 months prior to that. So this is a clue of index date misspecification, in that some of those measurement may be abnormal values suggesting prior disease. Another good clue for this is look at metformin. 8.8% of persons start metformin on day 0, but >10% had already started metformin in the year prior, so that suggests for those people, the first diagnosis date probably isnt the first date the patient was clinically recognized as having the disease (unless the person was taking metformin for some other reason).
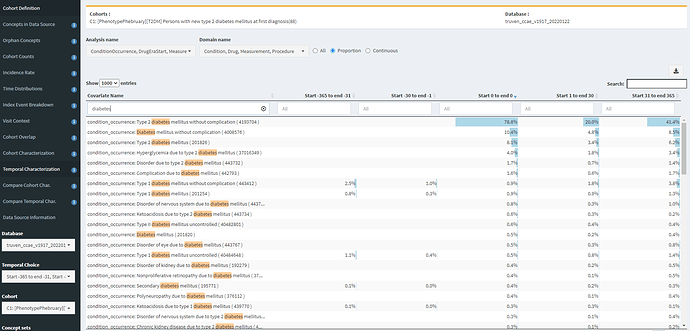
If I search for ‘diabetes’, I can also see examples of specificity errors:
Note, that ‘Type 1 diabetes mellitus without complication’ is observed in 2.5% of the persons in the year prior, 1% in the month prior, and 0.9% on the same day as the initial T2DM diagnosis. While the T1DM and secondary diabetes codes aren’t terribly prevalent, these explain the difference between definitions #1 and #2.
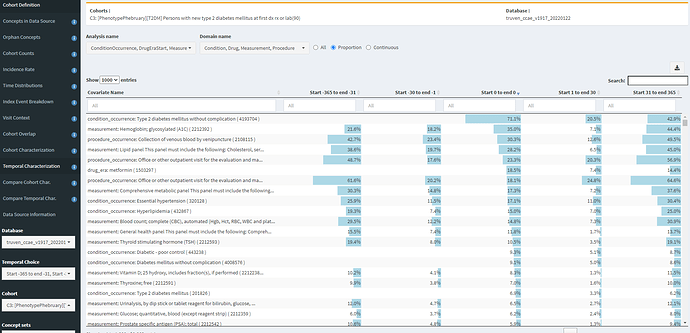
Now, in contrast, if we look at Temporal Characterization for definition #3 (indexed on diagnosis, drugs, or measurements):
We see that 18.5% of persons now have metfomin on day 0. We also see though that there remains a sizeable proportion of prior HbA1c measures. The CCAE database does not provide measurement values, only the record that a measurement was conducted, but this could be an indicator that still some residual index date misspecification persists.
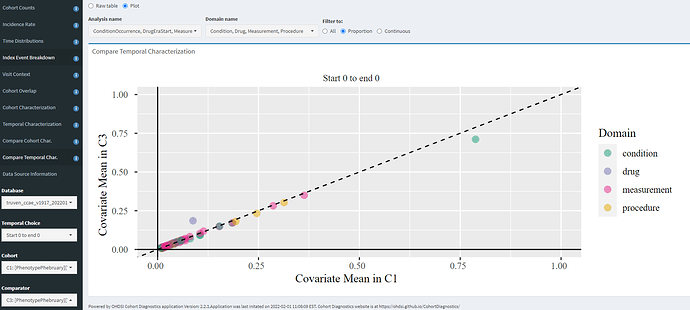
The ‘Compare Temporal Char’ tab allows you to evaluate two definitions head-to-head within a given database, for whatever features you may be interested in. Here, I’m zooming in only to the day 0 events to compare C1 (diagnosis only) vs. C3 (entry on diagnosis/drug/measurement), and we can see that - for this database- the only impact was an increase of day 0 metformin and corresponding day 0 decrease on ‘Type 2 diabetes mellitus without complication’. Otherwise, the patients looks very similar in characteristics (which should make sense, since the C3 cohort represents 80% of the C1 cohort).
Overall assessment
Based on this review, it seems there is clear index date misspecification associated with only using diagnosis codes, so I would recommend that a ‘new Type 2 diabetes’ cohort use additional information, such as drug exposure and measurement values to clean up prevalent cases. It also seems that there is some specificity errors associated with other forms of diabetes, so they can be successfully cleaned up using an approach similar to what is being used in LEGEND-T2DM, but they absolute impact of this appears low.
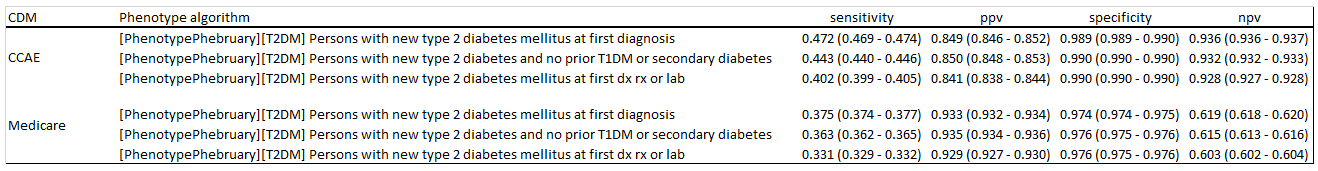
The six databases used here do not have complete measurement values, so the impact of measures is not fully understood at this point. We did not assess definitions based only on measurement values, which could potentially increase sensitivity (albeit with a likely negative consequence to specificity). I did not run PheValuator or adjudicate cases to try to estimate operating characteristics on any of these definitions, but that would seem like a reasonable thing to consider as a next step (and maybe something that someone would like to take on as part of this exercise).
So, hopefully this is enough information to get the conversation started. What do you all think? What have you learned about phenotyping Type 2 diabetes, from the prior literature or your own analyses? What insights did you gain by reviewing these definitions and the CohortDiagnostics results? Anyone interested in digging further into this evaluation? Share your thoughts, questions, comments, concerns, reflections, here on this thread, so we can all learn from each other.
Happy Phenotype Phebruary everyone!
 )
)