@Helen , you are definitely not too late (though I do appreciate that you’ve used a sloth as your icon for this post  ). Phenotype Phebruary isn’t stopping, we’re just using it to get the conversation started. You are register for access to atlas-phenotype by filling in this form: OHDSI Atlas Phenotype Library Registration . And also, I encourage you to join the Phenotype Development/Evaluation WG, which is run from within the OHDSI MSTeams environment, and you can sign up for this and any other WG you’d like here.
). Phenotype Phebruary isn’t stopping, we’re just using it to get the conversation started. You are register for access to atlas-phenotype by filling in this form: OHDSI Atlas Phenotype Library Registration . And also, I encourage you to join the Phenotype Development/Evaluation WG, which is run from within the OHDSI MSTeams environment, and you can sign up for this and any other WG you’d like here.
Thanks, Patrick. I already filled out the form last week but haven’t received an email about creating an account yet to access the resource. Do you have anyone to suggest to reach out to? Thanks ahead.
@Helen There are two different forms. One is to sign up for the Work Group, the other form (see link below) is for atlas-phenotype.ohdsi.org registration.
There is no record that you completed the form for atlas-phenotype.ohdsi.org registration.
Please use the link below to submit the atlas-phenotype.ohdsi.org registration form and your atlas account info will be mailed to you.
When I opened the link, it shows that I already filled the survey. Can you check it again? The email I used is hetinghelen@gmail.com, and I am from JHU SOM.
@Helen your Atlas Phenotype account has now been created and sent to you via email.
First of all, thank you for this impressive work! That was a joy to follow phenotype phebruary from the very first day.
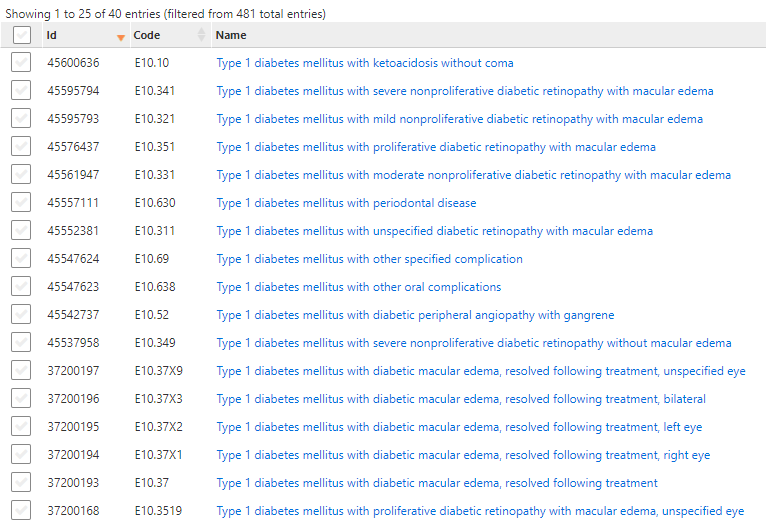
However, I think that there is an issue we have in this phenotype resulting from combination of Atlas behaviour and Vocabulary hierarchy. Using phenotype 1 with conditions only, I noticed that in Atlas included source codes, there are a few concepts with type 1 diabetes.
There are none of type 1 diabetes in included standard concepts, which suggests erroneous mapping. Lets take one code as example and see:
E10.10 Type 1 diabetes mellitus with ketoacidosis without coma is mapped to 2 conditions: 201254 Type 1 diabetes mellitus and 4009303 Diabetic ketoacidosis without coma, which is neither connected to Type 1, nor to type 2 diabetes, which is true, because any type of diabetes may result in diabetic ketoacidosis without coma. What is not true is that patients are included in Type 2 diabetes cohort despite explicit exlusion of Type 1 diabetes from the concept set. These codes were included by 442793 Complication due to diabetes mellitus and not excluded by 201254 Type 1 diabetes mellitus, which I think is a bit tricky of Atlas.
The pretty close issue is discussed right now in another topic.
We excluded Type 1 concepts by source code and the cohort was limited to only Type 2 DM patients.
So is it the expected behavior of Atlas and it just should be kept in mind or should be changed?
Am I missing something? Thoughts?
@Gowtham_Rao @Patrick_Ryan @Christian_Reich @Chris_Knoll @aostropolets @Alexdavv
Without knowing what the origin concept set expression is, I can’t comment on which concepts may be improperly mapped.
What I can say is that Atlas isn’t applying any special logic on top off the CDM vocabulary tables. When yous ay:
I’m not sure what trickery you’re referencing but when looking at the included source codes, you’re simply looking at the source codes that map over (via source-to-concept-map) to the standard concepts that are included from the concept set expression.
To put it another way (on how we get to included source concepts) we:
- Resolve the concept set expression into included conepts (uses CONCEPT_ANCESTOR)
- From the result of 1, find concepts via source_to_concept_map.
If the concept set expression wanted to include complications and excluded T1 concepts, then it makes sense that you got the complication included that happens to reference a T1DM cause…because the concept set expression is puling in any concept that’s a complication but the complication itself is not a T1DM diagnosis…
This is just some of the interesting side-effects you get from using the CDM vocabulary, and this is specifically why we give you the ‘included concepts’ and ‘included source code’ views in Atlas.
The likely conclusion here is that the vocabulary mapping is correct, and the concept set expression has to be more specific about which complications you want to find (such as only complications related to T2DM). But this is really a wild guess as I don’t know the specific code list and clinical concept you’re trying to model.
Thank you for the answer.
Type 2 diabetes, definition 1 has a concept set of conditions I am talking about. And I believe that concepts are mapped properly.
I also understand the mechanics behind it, what I found tricky is the side-effect. Anyway, the source codes referencing type 1 diabetes should be excluded from the mentioned definition.
If this effect is known, I just wanted to make sure that I am the only person considering it tricky ![]()
I played around with this, and I think that it’s a common trap where you think that if you include all disorders related to DM, but then exclude the ones related to T1, you would just be left with only those related to T2. However, as you saw, the 4009303 is something common to both T1 and T2 so you would get both source codes that indicate T1 and T2. So, IMO, using the ‘Complcation due to DM’ (ConceptID: 442793) is a little over broad because some of the descendants could be from Type 1 codes, and there’s nothign in the hierarchy that indicates that the Type 1 code falls under ‘complication due to Type 1 DM’.
I was able to remove the code by removing the general ‘complication due to DM’ and replaced it with ‘Complications due to Type 2 DM’, but I’m not sure what other side effects that may have, so I’m not suggesting anything related to the official T2DM phenotype. But this is a good excercise to show how using concept ancestor in the vocabulary may not always give you what you thought, usually because there’s a broader definition in the hierarchy that doesn’t have the type-specific children beneath it.
There is a concept for ketoacidosis due to type 1 diabetes (with a child that indicates ‘coma’) but there isn’t one that is specifically ‘without coma’…I don’t know if it makes more sense to map the E10.10 to 439770, but I’m pretty sure we got these mappings from SNOMED and so there was probably a reason to map to 4009303 vs 439770. But, that’s only half the puzzle because it doesn’t appear that 439770: Ketoacidosis due to type 1 DM roll up to ‘Disorder due to type 1 dm’, so it wouldn’t fully help us for purposes of how we are trying to define T2DM in this phenotype.
My 2 cents although with inflation it might not be worth what it use to be. You have to have the disease before you can have complications. How many people are entering a T1DM or T2DM cohort based solely on a complication code as the initial presenting diagnostis? Now yes it is very true that patient’s first experience with a condition may be as a result of a complication of said underlying disease which as of yet has not been properly recorded. I send my insurance company a quarterly HOMA-IR value so we will know precisely the first quarter upon which my beta cells starting deteriorating.
Nice to look back at this active discussion thread from Phenotype Phebruary 2022 
Just capturing the note that I’ll be using this cohort that’s in our OHDSI Phenotype Library as the basis for the nested indication for drug classes in our HowOften effort for OHDSI2023.
Interesting methodological issue: in the T2DM cohort, we correct for index date misspecification by allowing for indexing on drug or hbA1c measurement. However, in the context of nesting within the drug cohort, using the index date misspecification can induce ‘immortal time’, in that we ‘look forward’ from the hba1c measurement or drug to see the T2DM diagnosis, and the only circumstance that this would matter as a nesting cohort is if the measurement/drug is before the drug class index but the diagnosis is afterwards (and the time between the drug class start and the diagnosis is ‘immortal’ in that the person is guaranteed to survive until the diagnosis is observed). @jennareps has been worried about this problem in the context of patient-level prediction when defining target cohorts, but the problem is equally relavent to incidence characterization. It’s adjacent to the problem that @conovermitch and @jswerdel presented previously with immortal time due to ‘2 or more dx’ definitions. To address this in HowOften, I’m going to live with the index date misspecification and only use the diagnosis requirement.