Thank you @allanwu
atlas-demo.ohdsi.org has the following definitions - i have taken a snapshot of it.
Regarding Unaminity definitions:
Regarding output and shiny: I will update the shiny app output to reference the updated definitions in atlas-demo.ohdsi.org. this should have the attrition output. i.e. lets continue the iteration on atlas-demo.ohdsi.org and only move the mature definitions to atlas-phenotype.ohdsi.org at a future stage. This strategy will allow us to iterate and update quickly.
Regarding observations in the tiered definitions and how to address them: I suggest making a list of observations and frame them as sensitivity, specificity errors. You may also consider saying why you think they may be errors. This would help some of us non Parkinson’s experts provide ideas and collaborate on solving it. Examples are:
- variability in male and older preponderance by data source (e.g… possible sensitivity errors, younger individuals less likely to have Parkinson’s disease and mean age lower than expected)
- higher secondary parkinsonism (source of specificity errors?)
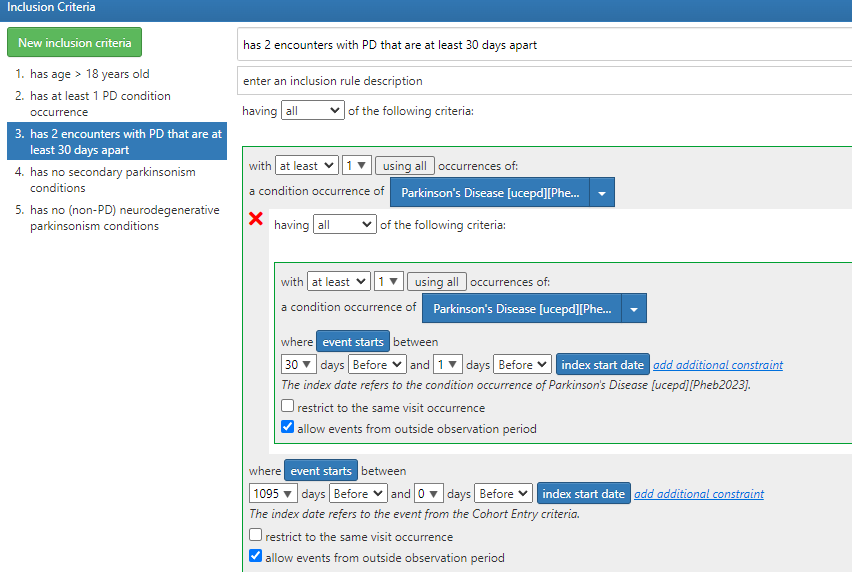
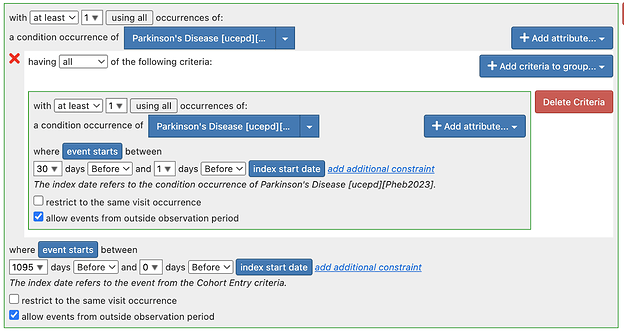
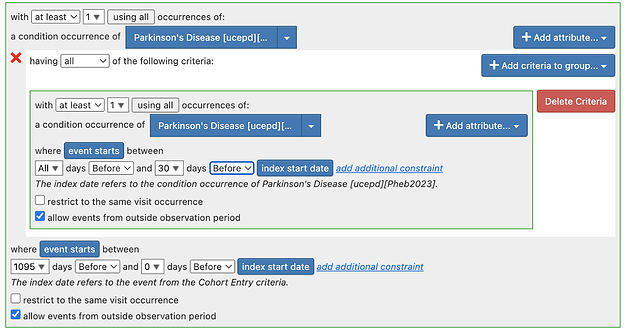
Regarding Tiered definitions: To pursue the tiered definitions, we need some time and it may well be atlas only, atlas + Phea or Phea only. But there are many outstanding issues in the atlas based definitions. Of note:
- Blanks for provider specialty: @Chris_Knoll shared with me that it is still a valid cohort definition and it will execute - but will make it more explicit
- There may be some logical errors in some of the definitions.
For next steps - for tiered definitions specifically - I think we should do a targeted re-review of the implemented cohort definition to make sure it is complete and has transformed the logic.
I am attempting to get an output on https://data.ohdsi.org/PhenotypePhebruary2023_P8_ParkinsonsDisease/ later today with updated output for at least the Unaminity definitions (it will have output of tiered definitions, but I think we should ignore them for now because of possible errors). I will update this thread when ready.