P8 topic for developing cohort defintions for Parkinson’s Disease
Phenotype defintion, adapted and updated from Phenotype Pheb 2022:
Phenotyping Parkinson’s disease (and neurodegenerative parkinsonism syndromes)
Allan Wu, MD. Revised 2/8/23
Goal: to develop and evaluate algorithms (and impact of criteria used within them) that assess incidence and prevalence of
- Parkinson’s disease (PD) and
- neurodegenerative parkinsonism syndromes (NPS)
(the NPS will not be part of Pheb2023)
Clinical description
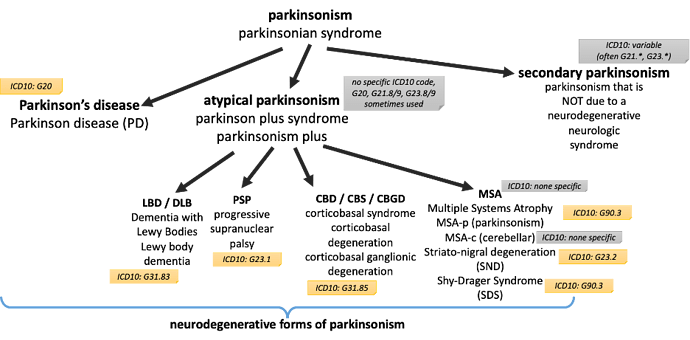
Parkinson’s disease (PD) is a neurodegenerative disease that has been estimated to be increasing in prevalence based on large scale epidemiologic work. It is the most frequent form of neurodegenerative parkinsonism, itself a subset of parkinsonism syndromes.
PD (also known as idiopathic Parkinson’s Disease) is the condition when there is specific degeneration of the substantia nigra dopaminergic (DA) neurons over time producing the disease. It is estimated that well over 50% of DA cells will degenerate before clinical symptoms appear. This is the classic and core definition; however, increasingly there is appreciation of that PD is a multisystem disorder with involvement of other neurotransmitters and other systems within and not within the nervous system (often noted as non-motor symptoms – constipation, cognitive impairment, sleep disorders, autonomic dysfunction, depression, anxiety etc).
PD is the most common form of neurodegenerative parkinsonism (80-85%).
Neurodegenerative parkinsonism syndromes (other than PD) are defined by having parkinsonism plus other neurologic systems that are affected by degeneration besides the specific PD degeneration described above. There will be described in more detail below – and include progressive supranuclear palsy (PSP), multiple systems atrophy (MSA), corticobasal degeneration (CBD), Dementia with Lewy Bodies (DLB), and others. Frequently, patients with neurodegenerative parkinsonism may be diagnosed with PD early on before neurodegenerative features become sufficiently prominent for a specific clinical diagnosis of NPS over time. If patients have a clear diagnosis of PSP or MSA (for example), those are not considered PD, but are within neurodegenerative parkinsonism.
A good reference and overview of PD as a clinical syndrome, its early diagnosis, and its evolution over time is Armstrong and Okun (2020 Diagnosis and Treatment of Parkinson Disease: A Review - PubMed)
Clinical diagnosis of Parkinson’s disease (PD)
Parkinson’s disease is a clinical diagnosis during life with no lab/imaging studies that definitively establish the diagnosis. Parkinson’s disease is the most common form of parkinsonism.
Parkinsonism is defined as the syndrome of
- rigidity
- rest tremor
- bradykinesia (slowness of movement)
- postural instability
Each of above can be recorded in a variety of different ways – and (except for rest tremor) tend to be highly nonspecific. The well-established practical UK Brain Bank criteria for parkinsonism is that 2 of the above 4 findings are present with at least 1 of the two being either rigidity or bradykinesia. Note, tremor is not required. The current Movement Disorders 2015 consensus criteria defines parkinsonism as bradykinesia with or without rest tremor, rigidity, or both – and must not be attributable to other causes (e.g. slowness from depression, drugs, illness, catatonia, lack of motivation, other neurologic disorders).
The current clinical diagnostic standard
Parkinson’s disease is clinically diagnosed when patients have parkinsonism and
- has supportive criteria without significant red flags or confounding findings.
Supportive criteria generally are accepted as - clear and dramatic beneficial response to dopaminergic medication – see below
- presence of levodopa-induced dyskinesia
- gradual progression over time
Red flags and confounding findings include - early (typically within 3 years of onset of sx) severe orthostatic hypotension
- early (within 3 years…) severe urinary incontinence/retention, not otherwise explained
- early recurrent falls (within first 3 years of sx)
- early severe dementia (controversial) – some criteria consider PD occurring independent of dementia; others will exclude early dementia or certain types of dementia
- complete absence of parkinsonism progression after 5 years
- neurologic findings such as ataxia, spasticity, aphasia, apraxia, supranuclear gaze palsy
- normal DATscan (dopamine transporter scan)
- absence of secondary causes for parkinsonism:
-
- drug-induced parkinsonism (typically has exposure to medication at the time parkinsonism is being diagnosed or considered and improves or does not progress if med is reduced, discontinued)
-
- multi-infarct vascular parkinsonism
-
- normal pressure hydrocephalus
-
- recurrent head trauma, severe traumatic brain injury or concussion with parkinsonism
-
- history of encephalitis with subsequent parkinsonism
The beneficial response to dopaminergic medication is considered highly specific, but a failure of response is difficult to prove as most criteria recommend 1000 mg/day levodopa be used without response before definitive criteria of not responding is registered. It is noted that this criteria makes having the prescription of a PD med not specific to making the diagnosis because it is not infrequently used as a “test” prescription to see if the patient will respond or not.
The progression of symptoms is slow and it may take several years before a clinically probable PD case is diagnosed. Quality measures for assessing quality care of neurologists have recommended annual re-review of diagnosis for the first 5 years after symptoms.
Conversely, many of the features that define neurodegenerative (non-PD) early in the course of disease (to be described below) become common in late-stage typical PD and one must be cautious not to overdiagnose neurodegenerative (non-PD) parkinsonism if it is clear that one has typical PD but that the patient has simply developed the other neurologic features as part of advanced late-stage PD disease.
Clinical diagnosis of neurodegenerative parkinsonisms (PD is one of them)
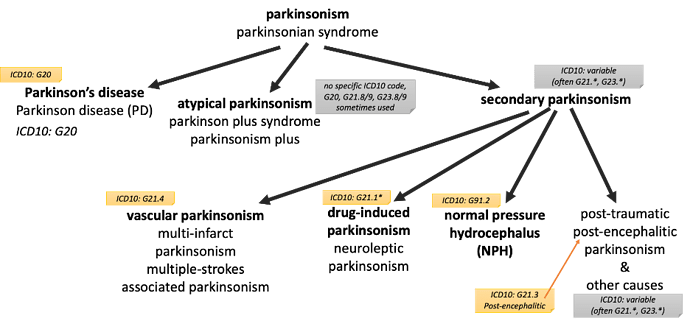
Non-neurodegenerative parkinsonism (secondary parkinsonism):
These should be excluded from cohorts of PD and neurodegenerative parkinsonism.
To distinguish neurodegenerative parkinsonism (of interest) from non-neurodegenerative parkinsonism can be challenging, especially since two categories may co-exist or be considered/coded early and over time, clinical clarity sometimes emerges.
Typically, the clinical diagnosis of a clearly established non-neurodegenerative parkinsonism which is correlated with the clinical course over time that makes sense for that particular etiology (varies by etiology) and which fully explains the neurologic presentation, then the likelihood of a neurodegenerative parkinsonism is then considered less likely.
Having a non-neurodegenerative parkinsonism (NNP) of course does not exclude PD (or other neurodegenerative parkinsonism) as the etiology may have unmasked an incipient PD which would be detected by abnormal clinical course that deviates from the NNP etiology over time.
These non-neurodegnerative parkinsonisms (by etiology) are often referred “secondary parkinsonisms” as exclusionary criteria. Common ones are vascular (multistroke) parkinsonism and drug-induced parkinsonism. Others described below.
Neurodegenerative parkinsonism (non-PD):
Among neurodegenerative parkinsonism, above clinical guidelines help distinguish PD from non-PD neurodegenerative parkinsonism.
The non-PD neurodegenerative parkinsonisms are defined by degeneration of other neurologic systems besides the specific PD pathology. Neurodegenerative parkinsonisms tend to progress over time, so time again becomes a distinguishing element as features below emerge gradually and can be nonspecific or not routinely assessed well enough to be difficult to recognize are present in an EHR.
Common neurodegenerative parkinsonisms (besides PD) and their associated symptoms / neurologic system involved:
-
PSP – progressive supranuclear palsy – parkinsonism + early falls & supranuclear gaze palsy
-
CBD – corticobasal degeneration – park-ism + asymmetric dystonia, apraxia, alien-limb
-
MSA – multiple systems atrophy – park-ism + autonomic dysfunction (bladder/orthostasis)
-
MSA-c – multiple systems atrophy – MSA + cerebellar ataxia
-
MSA-p – multiple systems atrophy – MSA + parkinsonism predominant subtype
-
SDS – Shy Drager Syndrome – MSA + predominant autonomic dysfunction
-
SND – striato-nigral degeneration – older term, not much use - considered parkinsonism without response to levodopa without other features above
-
LBD/DLB – parkinsonism + early dementia with fluctuating mental status, hallucinations and sensitivity to side effects of dopaminergic medications
-
LBD – Lewy body dementia or DLB (dementia with Lewy bodies) are considered part of the spectrum of parkinsonism – this is controversial, though for phenotyping neurodegenerative parkinsonism, it is reasonable to include within the “neurodegenerative parkinsonism” group that:
Often “atypical parkinsonism” or “parkinsonism” are used by neurologists to mean neurodegenerative parkinsonism though the specificity of the terms itself is often in question.
- “parkinsonism” is often used in documentation (and in coding) to identify the syndrome without committing oneself to the diagnosis of PD (either due to lack of findings, progression, and sometimes due to lack of experience or confidence in making the diagnosis). And unfortunately many EHR systems map “parkinsonism, not specified” to G20 (ICD10: Parkinson’s disease, primary parkinsonism), reducing confidence in G20 being as specific to PD as it could be.
A basic overview of these conditions are found in the same above PD reference as well:
Armstrong and Okun (2020 Diagnosis and Treatment of Parkinson Disease: A Review - PubMed )
Here is an overview provided by the Parkinson’s Foundation:
https://www.parkinson.org/pd-library/fact-sheets/parkinsonism-vs-parkinsons-disease
Phenotype development
Here’s the practical “clinician” oriented summary of the clinical diagnosis considerations above:
This hierarchy is not matched perfectly to SNOMED/OMOP-CDM hierarchy which follows.
- Parkinsonism syndrome
1.1. neurodegenerative parkinsonism (this is of interest #2)
1.1.1. Parkinson’s disease (this is of interest #1)
1.1.2. non-PD neurodegenerative parkinsonisms (aka “atypical parkinsonism”)
1.1.2.1 list of these: PSP, CBD, MSA (MSA-c, MSA-p, SDS), LBD, DLB, SND….
1.2. non-neurodegenerative parkinsonism (not of interest, but are often in the differential diagnosis under active consideration and if definitive and if the sole condition can be exclusionary); also known as “secondary parkinsonism”
1.2.1. examples: secondary parkinsonism – drug-induced parkinsonism, vascular/multiinfarct parkinsonism, normal pressure hydrocephalus, postconcussive parkinsonism, postencephalitic parkinsonism, etc
Literature on algorithms:
Review of many algorithms comes down to extraction of common elements with highly variable applications in several different combinations.
A good summary of 18 studies looking at algorithms for PD and parkinsonism (Harding et al 2019: Identifying Parkinson's disease and parkinsonism cases using routinely collected healthcare data: A systematic review - PubMed )
You can see a wide variety of PPV and sensitivity measures – and likely due to differing nature of cohorts, and as we would suspect, differing criteria for defining cases (PD or parkinsonism). Several of the broader ones also include secondary parkinsonism, mostly because there is a desire to capture broadly early cases of PD when it is often misdiagnosed or confused with secondary parkinsonism. Many used chart review as gold standard classification with some relying on neurology examination.
The CDC has created the National Neurologic Conditions Surveillance Survey (NNCSS) with PD as one of the first use cases (National Neurological Conditions Surveillance System (NNCSS) | CDC). In discussions with CDC (not published or available), a comprehensive review of algorithms seeking a case definition for PD revealed more than 120+ algorithms published. This is an opportunity to leverage the OMOP-CDM community to start the process of comparing algorithms in a uniform scalable way will be an important contribution. Our group is working to advise CDC on this process.
The elements that are most often used for PD / parkinsonism algorithms are as follows:
• Diagnosis conditions that support dx:
o Specific PD diagnosis
o Broader parkinsonism diagnoses
• Diagnosis conditions that represent competing or alternative diagnoses that, when are confirmed would exclude PD/neurodegenerative parkinsonism, but may co-exist or be an incorrect diagnosis early in course
• Diagnosis position (primary diagnosis or not for that encounter)
• Encounters (ambulatory only or ambulatory/inpatient/ED inclusive)
• Specialty coding the diagnosis condition
o Neurologist coding diagnosis conveys more confidence in diagnosis code
o Movement Disorder neurology subspecialty (not in OMOP-CDM vocab) conveys even further confidence in diagnosis coding
• Medications that support dx
o Specific PD dopaminergic medications (levodopa ingredient and dopamine agonists)
o Other PD medications fairly specific to PD (amantadine, COMT-inhibitors, MAO-B inhibitors, a few others)
• Symptoms and signs (rest tremor, gait imbalance, bradykinesia, rigidity) tend to be coded unreliably and uncommonly used in algorithms, though it remains a research questions about
All of above feature a look-back time (typically 1-5 years). This reflects the real-world chart review validation which starts with looking at diagnoses made most recently and by neurologists.
For diagnosis codes – typically # of coding events within lookback time as a threshold
For medications – either
- duration of meds must be sustained or
- count of prescriptions of meds over lookback time
One paper systematically evaluated different algorithms for detecting Parkinson’s disease with manual chart review as the adjudication standard (Szumski and Cheng 2009): Optimizing algorithms to identify Parkinson's disease cases within an administrative database - PubMed
Two algorithms were proposed that will be used for this P8 phenotyping exercise, summarized in table below. The tiered consensus algorithm has the highest overall accuracy (87% sensitivity with 83% PPV) and is dependent on an assessment of the expertise of the clinician that coded the diagnosis of PD. If a given patient was seen in movement disorders clinics, then the predominant code used by the movement disorders clinics was used to determine if PD was the final algorithm output (when compared with non-PD parkinsonism codes); if patient was seen only in non-movement neurology clinics, then predominant code used in those clinics would determine PD or not; if patient was never seen by a neurologist, then predominant code used in non-neurology clinics was used to determine PD or not. This algorithm supports the face validity that the specialist that codes the diagnosis has value over an algorithm that does not take into account the specialty.
The unanimity algorithm does not take into account the specialty and shows a lower overall accuracy, with lower sensitivity and PPV as in table below, though still improved over codes alone. Because these algorithms were developed using dataset gathered from a 3 year timeframe, we will also develop a phenotype using a 3 year lookback timeframe:
1.1. Tiered consensus accuracy 77%, sens 87%; spec 45%; PPV 83% Primary phenotype recommended; highest accuracy; good sensitivity; specificity is limited; not yet tested at population wide level; algo limited by specialty data availability
1.2 Unanimity algorithm Acc 67%, sens 78%, spec 33%, PPV 78% More complete data available; not dependent on specialty information present
1.3 Tiered consensus with medications Acc 75%, sens 77%, spec 68%; PPV 88% Meds increase specificity and PPV; allows assessment and identification of cases in primary algorithm who are not on meds (which is of exploratory interest)
1.4 Unanimity algorithm with medications Acc 64%, sens 60%, spec 75%, PPV 88% Adding medications adds specificity with some loss of sensitivity
PD: Primary phenotype for assessment:
Limitations – requires diagnosis codes
Differences from the published algorithm (Szumski and Cheng 2009)
• Lookback time of 3 years from last observation rather than fixed 3 year timeframe in paper
• Added a 30 day difference in 2 PD condition occurrences
• Includes all condition occurrences regardless of IP/OP (paper focused on OP)
• OMOP does not support movement disorder specialty
• Difficult to implement nested specialty criteria of neurologists
• And many changes for the tiered consensus algorithm will be needed.
1.1 tiered consensus algorithm (adapted Szumski and Cheng 2009)
• Initial event cohort:
o People having – condition occurrence of broad list of parkinsonism conditions
latest occurrence
• Inclusion 1: Age >= 18 yo at index date
• Inclusion 2: at least 1 occurrences of PD
o Event starts between 3 years before and 0 days before index start date
• Inclusion 3: at least 2 occurrences of PD separated by 30 days
o Condition PD starts at least 30 days before PD
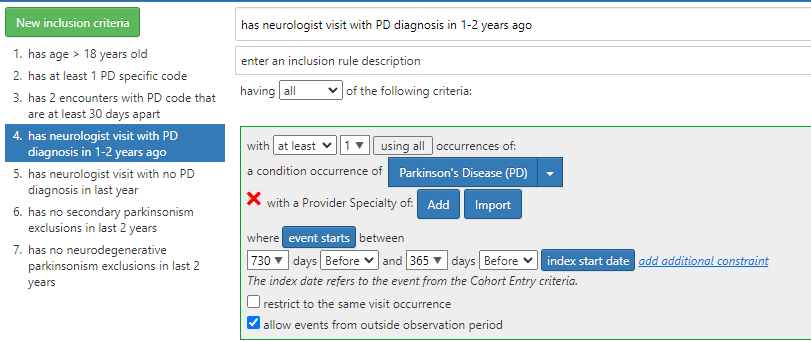
• Inclusion 4: Specialty criteria
o This will be challenging to implement; bottom line is to come up with a way to represent tiered specialty dependent criteria – perhaps “PD condition coded in all visits in the last 1 year of observation”
Visit occurrence with two levels of provider specialty
• “Movement Disorders” (this does not exist in the NUCC and is not an accepted standard specialty), though in common clinical use
o Likely not use in initial OMOP definition
o Could leave a placeholder criteria here so that individual sites can add local location-or-provider ways of specifying what is a “Movement Disorders” specialty visit.
• “Neurologist” neurology, neuro-opththalmology, neuromuscular medicine, neurology with special qual in child neurology, clinical neurophysiology, vascular neurology, neuropsychiatry, clinical neurophysiology by NUCC criteria
o Nested criteria (lookback 3 years for all criteria below):
o If person has at least one visit with provider specialty with “movement disorders”
If PD Condition code is the most frequent condition coded, compared to condition codes for non-PD parkinsonism conditions, then this criteria is met (true), else…
o Elseif person has at least one visit with provider specialty with “neurology”,
If PD Condition code is the most frequent condition coded, compared to condition codes for non-PD parkinsonism conditions, then this criteria is met (True), else…
o Else (patient was never seen by neurologists)
If PD Condition code is the most frequent condition coded, compared to condition codes for non-PD parkinsonism conditions, then this criteria is met
•
Inclusion 4: Specialty criteria logic structure for Atlas (2/8/23 11:30am)
o A PD condition exists in last 3 years - applies to all below criteria
o If there exists a Visit with Neurology specialty (OMOP Visit Provider specialty)
If PD condition exists with a Neurology Visit in the last 1 year,
• If there are no exclusion parkinsonism dx by Neurology visits in the last 1 year,
• Then, criteria is met
Elseif PD condition exists with Neurology Visit 1-2 years before index event,
• If there are no exclusion parkinsonism dx by Neurology visits 0-2 years before index event,
• Then criteria I met
Elseif PD condition exists with Neurology Visit 2-3 years before index event,
• If there are no exclusion parkinsonism dx by Neurology visits 0-3 years before index event,
• Then, criteria is met
Else (no PD condition exists with Neurology within 3 years),
• <follow same logic as “No Visit with Neurology below>
End
o Else (there does not exist a Visit with Neurology specialty in last 3 years),
“No Visit with Neurology” logic:
If PD condition exists within the last 1 year of observation,
• If there are no exclusion parkinsonism dx in the last year,
• Then criteria is met
Elseif PD condition exists 1-2 years before index event,
• If there are no exclusion parkinsonism dx in 0-2 years before the index event,
• Then criteria is met
Elseif PD condition exists 2-3 years before index event,
• If there are no exclusion parkinsonism dx in 0-3 years before the index event,
• Then criteria is met
Else
• Criteria is NOT MET
End
o End
1.2 Unanimity algorithm (adapted Szumski and Cheng 2009)
• Initial event cohort:
o People having – condition occurrence of broad list of parkinsonism conditions
• Inclusion 1: Age >= 18 yo at index date
• Inclusion 2: at least 1 occurrences of PD
o Event starts between 3 years before and 0 days before index start date
• Inclusion 3: at least 2 occurrences of PD separated by 30 days
o Condition PD starts at least 30 days before PD
• Inclusion 4: No competing condition codes
o between 3 years before and 0 days before index:
o Condition codes related to neurodegenerative (non-PD) parkinsonism
o Condition codes related to secondary parkinsonism
1.3 and 1.4. adding medication criteria (Szumski Cheng 2009)
At least one medication for PD (“PD specific medication list”)
Inclusion diagnoses:
ICD code for PD: ‘G20’, ‘332.0’
ICD codes related to neurodegenerative parkinsonism includes ‘G23.1’, ‘333.0’, ‘G31.83’, ‘331.82’, ‘G23.9’, ‘331.6’, ‘G90.3’,‘G23.2’, ‘G23.3’,‘G31.85’
The ICD codes related to red flags (confounder codes) includes ‘F20’, ‘F20.0’, ‘F20.1’, ‘F20.2’, ‘F20.3’, ‘F20.5’, ‘F20.8’, ‘F20.81’, ‘F20.89’, ‘F20.9’, ‘295’, ‘295.0’, ‘295.00’, ‘295.01’, ‘295.02’, ‘295.03’, ‘295.04’, ‘295.05’, ‘295.1’, ‘295.10’, ‘295.11’, ‘295.12’, ‘295.13’, ‘295.14’, ‘295.15’, ‘295.2’, ‘295.20’, ‘295.21’, ‘295.22’, ‘295.23’, ‘295.24’, ‘295.25’, ‘295.3’, ‘295.30’, ‘295.31’, ‘295.32’, ‘295.33’, ‘295.34’, ‘295.35’, ‘295.4’, ‘295.40’, ‘295.41’,‘295.42’, ‘295.43’, ‘295.44’, ‘295.45’, ‘295.5’, ‘295.50’, ‘295.51’, ‘295.52’, ‘295.53’, ‘295.54’, ‘295.55’ , ‘295.6’, ‘295.60’, ‘295.61’, ‘295.62’, ‘295.63’, ‘295.64’, ‘295.65’, ‘295.8’, ‘295.80’, ‘295.81’, ‘295.82’, ‘295.83’, ‘295.84’, ‘295.85’, ‘295.9’, ‘295.90’, ‘295.91’, ‘295.92’, ‘295.93’, ‘295.94’, ‘295.95’, ‘F25’, ‘F25.0’, ‘F25.1’, ‘F25.8’, ‘F25.9’, ‘295.7’, ‘295.70’, ‘295.71’, ‘295.72’, ‘295.73’, ‘295.74’, ‘295.75’, ‘F21’, ‘301.22’, ‘G24.1’, ‘G24.2’, ‘G24.3’, ‘G24.4’, ‘G24.5’, ‘333.6’
Confounder codes related to secondary parkinsonism includes ‘A52.19’, ‘094.82’, ‘G21’, ‘G21.0’, ‘G21.11’, ‘G21.19’, ‘G21.2’, ‘G21.3’, ‘G21.4’, ‘G21.8’, ‘G21.9’, ‘332.1’, ‘G91.2’, ‘331.5’
PD specific medication list: any med with levodopa ingredient (typically forms of carbidopa/levodopa); pramipexole, ropiniorole, rasagiline, rotigotine, apomorphine, entacapone
SUMMARY of discussion today 2/8/23 am.
Thank you to all participants to our 1 hour discussion to help with developing our cohort defintion. @Gowtham_Rao @Azza_Shoaibi @fabkury and others (sorry didn’t catch everyone who attended)
We agreed to prioritize the unanimity algorithm with and without meds.
These were tweaked in real-time on the Atlas-Demo site (since my local instance of OMOP-CDM is not yet up and running):
The Unaminity phenotypes are on Atlas-Demo, tag [Pheb2023]
- ID: 1781748
- ID: 1781760
Paper referencing these algorithms: Optimizing algorithms to identify Parkinson's disease cases within an administrative database - PubMed
OHDSI team will help clean up/review logic of the Atlas Demo cohorts and check the concept sets in readiness for a network study.
The tiered consensus logic is challenging to adapt. The cohort definition post included a outline of how this COULD be implemented in Atlas. It is also challenging to use specialty as a criterion when we suspect different OMOP-CDM instances may vary in how this is loaded/coded.
I believe it is worth attempting an implemenation of the tiered consensus algorithm (a) because it has a promise of improved sensitivity, specificity, PPV and (b) may help identify gaps in OMOP-CDM implementations.
Also, need to consider the Phea SQL builder for more complex logic that tiered consensus algorithm may be able to utilize.
Thanks for the great work, @allanwu. The formatting got broken but I am glad to be able to see all the details of the logic and codes, including your version of inclusion 4 in Atlas.
Another awe-inspiring detailed work. Sorry I couldn’t be in the meeting. Couple questions:
- Looks like the algorithms studied are trying to distinguish Parkinson’s disease from its “mimics”, as the paper calls them, correct? Or are we also worried about complete non-cases?
- I am not following what the thing is we are trying to catch. Is it PD only or PD + neurodegenerative parkinsonism?
- What are “confounder codes”? Those of the mimics?
- You say the algorithms are optimized for incidence and prevalence rates. I guess, that is in the general population, right? We are not talking outcomes? Can it be outcomes? PD is idiopathic, so by definition it cannot be the result of some “cause”.
- If it is not outcomes, but a patient population to study, say, the effectiveness or safety of drugs: Do we need a highly complex algorithm, which is hard to build even in US data, leave alone overseas? Or aren’t patients exposed to one or the other drug sufficiently selecting the right target?
- The algorithms in the literature show a low specificity (unless combined with medications, which renders them infeasible for many questions such as testing for treatments not included in the list). What are these patients? The mimics, or just random unrelated stuff? If it is the former - aren’t the vascular/drug related mimics much less frequently? How can they sneak into the cohort in such large numbers? If it is the latter, how come they are coded as PD? In other words, what demon are we fighting here? Do we know?
I stand by my general critique: We need to modularized bottom up algorithms, starting with the diagnosis code and explicitly calling out each inclusion criterion for its purpose, and how it does in the CohortDiagnostic/Phevaluation.
Thank you @allanwu, for this explicit discussion about PD, in addition to the medication list that you provided, do not you think “amantadine” is also be included especially for those with early PD? Amantadine and Rimantadine are antivirals used to treat Influenza A and B. Amantadine was used for patients with early PD symptoms.
Thanks for the input! It would be of interest to include amantadine or not.
We elected to stay with the general classes that were described in the paper and tried to keep it to the more specific medications used for PD.
So, we left out anticholinergics for this reason.
It would be reasonable to include amantadine/rimantadine, particularly since there is some pharmaceutical development on long-acting amantadine marketed toward PD.
But for this first pass of PD cohort efforts, we kept the concept set for PD meds closer to what was published.
Once we get through this front to back cohort definition, evaluation, publishing, we have many variations of these criteria we want to evaluate as well.
Thanks Christian, it is wonderful to participate with this group more actively.
I can provide a bit more context to the relatively broad phenotype descriptions and the focus now on PD itself.
-
The paper and the focus of this evaluation P8 exercise is to identify classic, typical, idiopathic PD (the problem is indeed so many ways to describe it) and, in this concept, PD is different from the non-PD neurodegenerative syndromes that are distinct clinical entities. So a PSP or atypical parkinsonism is indeed coded as not-PD for our desired cohort.
-
The bigger picture context is that we are advocating for state-wide and population-wide surveillance registries (California, Nebraska, CDC) that are gaining momentum to be more (not less) inclusive of possible parkinsonism b/c of the overlap of clinical symptoms/signs among PD and mimics (secondary parkinsonism and non-PD neurodegenerative parkinsonism). Many of these share epidemiologic risks, healthcare demand/burden, and the confusion of diagnoses too early (and interest in identifying them early) motivate being more senstiive, less specific (to the point of being acceptable to stakeholders who are reporting).
Once the PD surveillance registries are established, there is need to “parse” these public health datasets into more useful subcategories. This is where being as reasonably specific to PD is. Later work will create the other useful subcategories such as “too early” and each individual non-PD parkinsonism syndrome (PSP, MSA, CBD, etc).
so bottom line, is this cohort defintion is PD as an important P8 example, but the phenotype was written more broadly b/c of the need to capture the full context.
-
what are “confounder” codes. Those are legacy codes not used in the paper and the current proposed Atlas cohort defintions. Those were mostly diagnoses such as schizophrenia, other psychiatric, traumatic brain injury conditions that would be at high risk for getting medications or having injuries that would produce secondary parkinsonism. In the end, these were included b/c they were part of an intiial highly specific cohort but should have been removed from current efforts. They were not fully justified by literature.
-
Question about incidence and prevalence rates. Indeed, the motivation for our work is to improve the population-wide estimates of incidence and prevalence given momentum in California and the CDC for creating a surveillance structure for PD (and we advocate for NPS parkinsonism as well). The emphasis (and driver of the early registries, particualrly California) was epidemiology work. So that is why we are more concerned about sensitive capture of a potential population and cohort subset of specific PD.
Yes, as a neurologist, we are indeed interested in outcomes and driving improved care. The leaders of our group, including myself, are involved with the American Academy of Neurology Axon Quality registry, which includes a number of quality measure for PD. They are overall poor and a whole other topic b/c they overemphasize, in my opinion, process measures (screen for autonomic dysfucntion once a year) rather than true outcomes (which the field has yet to come to practical conesnsus on (i.e. ED visits for falls + broke bones tends to be what is used).
-
good point to study effectiveness/safety of drugs. More than drugs, i think the field of PD is more interested in precision medicine with subtypes of PD that respond differently to treatments like drugs. We’ll have to tackle this once we have our proof of principle OMOP-CDM cohort efforts (and we get our own OMOP-CDM instance up and running).
-
Super great questions. What are all these parkinsonisms that are not PD? Indeed we are ok with lower specificity at this point. We want to encourage surveillance registires (as a first step toward interoperable state-wide/population wide PD registries at scale) for completing epidemiology and public health work. But what do you do if you have such data? I’m a neurologist so I want to know what subset of this population data actually have some specificity for PD (current work) to drive improved care of healthcare distribution. The epidemiologists (and the patient advocates) want to know the incidence/prevalnce of the most sensitive cases (to advocate for more funding and emphasize how many “parkinsonisms” there are.
I will say one final comment about the challenges here. IMO and other EHR mappings commonly map “parkinsonism, NOS” as G20 (which represents Parkinson’s Disease". This is not just the fault of IMO and mappings, but is confounded at the level of ICD10 defintion as well. This automatically makes the G20 code (ICD10 for PD) – mapping to SNOMED/OMOP-CDM standard – as less than ideal to identify PD.
@allanwu as a follow-up to the discussion referenced above
this is an executable R package that may be executed as part of OHDSI-studies network study GitHub - ohdsi-studies/PhenotypeParkinsonsDisease
I have executed this package on a few data source and the output is available below. I think the next step is to make some initial inferences on its performance characteristics. You may also use the output to check if there are logical errors in Cohort definition (ie if something looks unexpected, then probably there was some error someplace in our atlas definition design).
https://data.ohdsi.org/PhenotypePhebruary2023_P8_ParkinsonsDisease/ .
As you described above, this only has the Unanimity algorithm based cohort definitions. Tiered based cohort definitions need to be developed. I understand the main operating characteristic of interest is specificity and positive predictive value. So I think it’s reasonable to look at the population level summary characteristic in the tool to check for anything that looks unexpected and would be evidence that there are person’s in the Cohort that don’t have the phenotype of interest. As we learn, it is reasonable to iterate and improve the definition, if needed.
This is an interesting design and it is different from almost all definitions I have worked on before. Reason, it’s indexed on last encounter (index date) apply rules to improve specificity on time temporally prior to index date.
Looking forward to learning with you.
Thanks @allanwu for driving this conversation, and thanks also @Gowtham_Rao for implementing the discussion and providing summary data for the community to review and learn from together.
One thing that I was wondering about in the cohort definition implementation: it appears these cohort definitions are indexing off the latest of the parkinsonism concepts and then looking back to see if persons have 2+ PD codes and no secondary or neurodegenerative codes. But the consequence of this definition is that a person’s cohort_start_date will be the latest point they were observed. Wouldn’t we consider a person to have Parkinson’s at the first date of diagnosis, so long as they satisfy the same criteria (>=1 code >30d apart and no secondary or neurodegenerative codes before or after)? Or is it specifically the case that once a secondary or neurodegenerative diagnosis is recorded, a person is reclassified out of the PD cohort (which means you’d have to ‘look forward’ from the initial PD diagnosis date to rule out future reclassification)?
I created a new cohort definition (JSON attached here) that tries to ‘reindex’ based on when the disease was first diagnosed. Interestingly, I couldn’t get the exact same patient count as the original cohort (it was very close, but not exactly the same), and it’s because theres a small proportion (<0.1%) who have a secondary or neurodegenerative concept AFTER the latest PD code.
PD reindex.txt (14.7 KB)
Addressing @Patrick_Ryan post, the issue is PD is not diagnosed either early or late. Patients are diagnosed with various parkinsonisms nonspecifically for several visits b/c it is too subtle or the provider remains unsure of the diagnosis. Later, it is often clearly reasonably coded/diagnosed as PD (even though it is actually a neurodegenerative parkinsonism),. Then later visits are most accurately diagnosing the person with either PD (if it does not change for many years) or with a PSP/CBD/MSA parkinsonism. So the most accurate diagnosis is usually considered the most recent “set” of most consistent diagnoses (usually neurologists) b/c the primary care docs often just keep copying forward the erroneous PD diagnosis code even if the neurologist has diagnosed something differently later.
This is supported by the note that the <0.1% proportion have a secondary/neurodegenerative concept AFTER the last PD code.
We are intensely interested in the earliest entry/incidence date in our cohort, but to classify if the person is actually PD or Not PD requires look at the last several visits (and we suspect specialty, but that’s to be tested). The literature suggests that once you figure out someone has PD later, then the earliest “broad parkinsonism” concept counts as start of cohort which is how Atlas seems to work. I do lose this in the way @Gowtham_Rao and I constructed the unaminity algorithm.
I tried to copy/paste JSON in the post into Atlas-demo and I didn’t see a difference in the indexing of cohort start. See cohort 1781774:
Thank you @Gowtham_Rao for sharing the initial characteristcs across 12 databases. In my first pass, I have a few observations. I hope the team here does not mind my feedback on use of tool mixed in here.
- Comparing the cohorts without and with med criteria allows us to see how much variability there is between databases in defining PD using a med criteria or not. I computed a % of dropoff in counts from the PD without a med criteria vs PD with a med criteria.
These vary from 8% to 79% across 12 databases, likely reflecting how each database robustly (or not) represents medications. The overall dropoff is 33% (from 839,007 counts w/o meds to 561,482 w/meds. Interestingly, our literature paper showed a dropoff from 387 to 368 (4.9%) - lower than any of our OMOP databases.
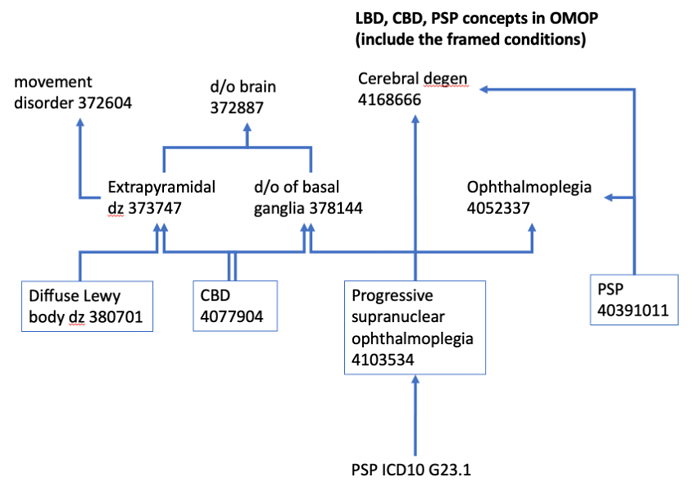
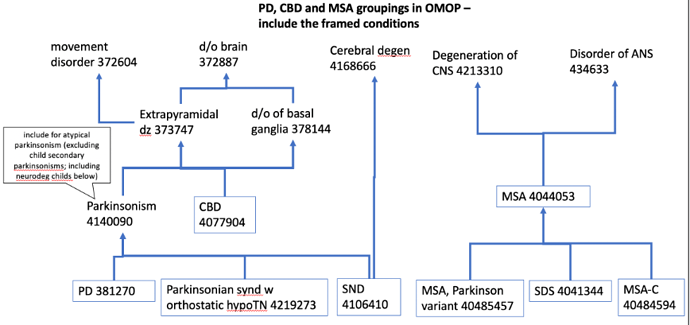
Orphan concept picked up the need to ADD:
concept 4103534 progressive supranuclear ophthalmoplegia to the ‘parkinsonism (non-PD, neurodegenerative) conditions’ concept set.
@Gowtham_Rao can you make this change in the two unanimity packages?
I’ve updated it in Atlas-demo already.
Nice feature!
Couldn’t run incidence check: "Error: there is no package called ‘ggh4x’ "
In general, have to be careful reading the help text esp since these cohort defns are based on most recent observation rather than earliest… For example, How to interpret cohort start and end when entry point is “most recent” event?
Visit context useful – most databases were all outpatient.
The claims and pharmacy ones were much more heterogeneous. Likely accounts for some variation.
(it would be nice to see associated specialties with visits - but realize this is not yet standard in OMOP-CDM).
cohort overlap function to create table did not work (good graphs though).
Most cohorts had expected characteristics – male predominance.
- the ages appear high but likely b/c our index event is “most recent”
- associated events make sense - depression ,dementia make expected appearances; sleep disorders interestingly did not show up much at all - REM Behavior Disorder is something I was looking for and is a prodromal syndrome in PD/parkinsonisms
- would be nice in the pretty version to be able to filter in/out classes of concepts. I was looking to focus on neuropsychiatric symptom/syndrome conditions and exclude some of the general medical geriatric conditions (hypertension, renal dz, etc).
Two databases show unusual characteristics, younger and F>M
- these seem to have higher rates of concepts of secondary parkinsonism than the others (truven_mdcd, jmdc databases)
- the fact these show up in our unanimity cohort suggest that either the source EHR/ETL maps ‘vague parkinsonism’ to ICD10 G20 → which becomes PD OR we are missing a secondary parkinsonism in our exclusion criteria somehow.
- to figure this out, I would want to look more closely at the younger age female cohorts associated condition/concept characteristics in these 2 databases.
Proposed Atlas PD tiered consensus algorithm.
@Gowtham_Rao if time, can we have this logic reviewed?
Will be a great comparison to unaminity algorithm.
Atlas-demo did not have ability for me to populate Specialty – so those are blank but intended for “neurology” specialty.
I think I have worked out logic adapting the tiered consensus algorithm to Atlas.
Basically, because Atlas and OMOP cohorts work by successively criteria that narrow counts, it struggles with logic that has to do with multiple and sequential criteria (i.e. specialty and most recent dx)
So I set up 6 Atlas cohorts, each of which is designed to be based on non-overlapping distinct subcohorts. By assembling output from all 6, we achieve the counts needed for this algorithm.
Logic:
Identify non-overlapping buckets. Each will be its own Atlas cohort
All have two PD conditions in the last 3 years (no restrictions by visit type/specialty)
Looks like 6 buckets - each with subset of what would fulfill criteria
- neurologist/PD in last year
- neurologist/PD in 1-2 years ago (not last year)
- neurologist/PD in 2-3 years ago (not last 2 years)
- non-neuro/PD dx in last year (never saw neurologist in 3 years)
- non-neuro/PD dx in 1-2 years ago (not last year)
- non-neuro/PD dx 2-3 years ago (not last 2 years)
neuroPD1year cohort - 1781732
Has seen neurologist in last 3 years (redundant do not implement)
Has PD condition by neurologist in last 3 years (redundant do not implement)
Has PD condition by neurologist in the last year
Has No exclusion conditions by neurologist in last year
neuroPD2years: cohort 1781779
Has seen neurologist in last 3 years (redundant)
Has PD condition by neurologist in last 3 years (redundant)
Has PD condition by neurologist in 1-2 years ago and NOT in last year
Has No exclusion conditions by neurologist visits in last 2 years
neuroPD3years: cohort 1781780
Has seen neurologist in last 3 years (redundant)
Has PD condition by neurologist in last 3 years (redundant)
Has PD condition by neurologist in 2-3 years ago and NOT in last 2 years
Has No exclusion conditions by neurologist visits in last 3 years
neuro-noPD (not a cohort - here for logic completion)
Has seen neurologist in last 3 years
Has NO PD condition by neurologist in last 3 years
Does not fulfill criteria for cohort
Can infer counts for this based on above 3 cohort exploration
nonneuroPD1year: cohort 4 of 6: 1781781
Has not seen neurologist in last 3 years
Has PD condition by visit in the last 1 year
Has No exclusion conditions in last year
nonneuroPD2year: cohort 5 of 6: 1781782
Has not seen neurologist in last 3 years
Has PD condition by visit 1-2 years ago and NOT in the last 1 year
Has No exclusion conditions in last 2 years
nonneuroPD3year: cohort 6 of 6, cohort 1781783
Has not seen neurologist in last 3 years
Has PD condition by visit 2-3 years ago and NOT in the last 2 years
Has No exclusion conditions in last 3 years
nonneuro-noPD (not a cohort; here for logic completion)
Has not seen neurologist in last 3 years
Has no PD condition with a visit in last 3 years
DOES NOT MEET CRITERIA
Can infer counts from above 3 cohorts
Great conversation.
1. Regarding re-indexing: @Patrick_Ryan regarding re-indexing - in the video recording (see link) we discussed this. @allanwu made a point that the operating characteristic he is looking for in current iteration was specificity and PPV. Index date misspecification error was acceptable at this time. Also the intent was to replicate a published algorithm, which was indexing on latest event. In the group discussion, it was decided to explore reindexing after achieving these above goals.
2. Regarding reindexed cohort definition developed by @Patrick_Ryan : Btw @allanwu i fixed this
3. Regarding comparing Tiered definition with unaminity algorithm:
This is purely technical - we cannot execute a cohort definition when the cohort definition is incomplete i.e. provider specialty. i.e. i cannot re build cohort definitions without this issue being fixed
4. Regarding bugs in software at data.ohdsi.org/PhenotypeLibrary . The issue appears to the server infrastructure running the OHDSI tool. The error in incidence rate plot (tagging @lee_evans and @jpegilbert has been previously reported but I think there were some challenges in fixing it
@fabkury - I can’t seem to find your post now which I saw this morning where you added a Phea SQL tweak to a placeholder in Atlas-demo for the tiered consensus.
Two points to make on the logic as I recall. You are counting the # of PD and non-PD (neurodegenerative) parkinsonisms in each visit and only considering visits where # of PD conditions are greater than the other parkinsonism exclusions.
- The tiered consensus criteria takes all visits in the 3 year time frame and counts all conditions PD and (non-PD parkinsonism exclusions) in total across all visits. Then the tiered consensus algorithm/function outputs “PD” if the PD conditions outnumber the (non-PD parkinsonism exclusions).
- The (non-PD parkinsonism exclusions) should be the combination of (non-PD neurodegenerative parkinsonisms) and (secondary parkinsonisms) concept sets.
@allanwu, yes on both items 1 and 2. Thanks for revising my message.
Here is a summary of the “Phea approach” for the tiered consensus algorithm for Parkinson’s Disease (adapted Szumski and Cheng 2009, see @allanwu’s message above).
What is the tiered PD phenotype, and what’s the matter with it?
The tiered consensus algorithm phenotype requires PD to be coded more frequently than competing diagnoses, within the past 3 years. The competing diagnoses are non-PD parkinsonism and secondary parkinsonism.
The problem is that Atlas can’t do such a “most frequent diagnosis among 2 groups in the past 3 years” calculation. In inclusion 4 above, @allanwu tried to approach that logic in Atlas’s Cohort Definitions tool by scanning the prior 3 years one year at a time.
Goal
Create a phenotype that correctly applies the tiered logic (“most frequent among 2 groups” logic). Do that by injecting Phea-generated SQL into an OHDSI-compatible cohort definition (cohortDefinitionSet).
Whether the visit is a neurology visit or not, was not considered. This aspect of the logic was ignored because we thought it would be too difficult and unreliable to map “neurology visit” across sites in a network study.
Logic to be computed
Tiered diagnosis criteria: Parkinson’s Disease is more frequent than competing diagnoses (non-PD and secondary parkinsonism).
Query logic:
- At every visit occurrence, look back and count the number of occurrences of the two groups of conditions, PD and non-PD.
- Eliminate the visits where the non-PD count is bigger than the PD count.
- Patient meets the criteria if it has at least one of those “special” visits that remain.
Alternative logic (not used): Use most recent diagnosis, instead of most popular.
The hard part isn’t producing a SQL query that captures the above logic (and you have Phea to write that query for you). The hard part is correctly plugging in that “special SQL” code into the OHDSI’s ecosystem, i.e. into an OHDSI study package.
Challenges
-
Phea SQL needs to be compatible with the local SQL flavor. (not yet addressed, Postgres was used)
Possible solution A: have the study package generate Phea SQL locally. -
Phea SQL needs to be compatible with CohortGenerator / OHDSI-SQL.
Possible solution A: replace a placeholder criterion with Phea SQL. (this was the approach taken)
Possible solution B: have Phea insert rows into@target_database_schema.@target_cohort_tabledirectly, without going throughCohortGenerator. -
I can’t test the code post-Phea, because neither Synthea nor Eunomia have any Parkinson’s disease diagnosis code.
Potential solution: For testing purposes, surrogate conditions could be used. (this was not done)
How the “SQL replacement approach” works
-
Manually copy the cohort definition created by the group (https://data.ohdsi.org/PhenotypePhebruary2023_P8_ParkinsonsDisease/) into a new one (ATLAS).
-
Manually add an extra criterion in that copy:
has visit where PD Dx is the most frequent.
(subsequent steps are done by R code, see TXT file attached)
-
Download the cohort definition using
ROhdsiWebApi::exportCohortDefinitionSet(). -
Read the SQL file that was downloaded. Replace the extra criterion with the Phea SQL.
a. Before this substitution, adapt Phea’s code to be compatible with OHDSI-SQL:
i. Replace schema references with OHDSI aliases (e.g.@cdm_database_schema).
ii. Retrieve code sets from the temporary table#Codesets. -
Generate a new study package using the modified SQL file. (did I do this correctly? I am not sure)
The modified study package is in MS Teams:
- Modified cohort definition: cohort 1781786 Phea.zip
- Modified study package: PhenotypeParkinsonsDisease-phea.zip
Comments and takeaways
-
Local generation of Phea SQL (at data holder’s computer) is complicated by:
a. Phea needs to read the column names to build the SQL query. But Phea doesn’t know the details about the local CDM like SqlRender does. Therefore, OHDSI-SQL aliases need to be resolved into real table names and provided to Phea. -
Probably can’t edit the cohort definition after the SQL replacement. It is likely that you will lose Phea’s SQL code when the cohort generation SQL gets re-generated.
-
Alternative approach: Use Phea to directly insert rows into
@target_database_schema.@target_cohort_table, with or without going through CohortGenerator. -
Long term best solution: Develop Phea all the way into a HADES-integrated R package.
Files in MS Teams’ Phenotyping Development Workgroup team
- Edited cohort definition: cohort 1781786 Phea.zip
- Edited study package: PhenotypeParkinsonsDisease-phea.zip
R code that generates the Phea SQL and performs the “SQL replacement” pipeline
parkinsons.txt (8.2 KB)
Final words
If anyone trusts me enough to download PhenotypeParkinsonsDisease-phea.zip from MS Teams and run the study on your local CDM instance, that would be nice! Take a look at file 310.sql if you want to see the edits that were applied (search for keyword “phea” in the file). But I admit the chance that it will work is very small. That is because, while I could test the query logic per se in my local Postgres server, I don’t have a fully-featured local OHDSI environment for running the study package. Moreover, the code was generated for PostgreSQL – maybe it happens to work in other SQL flavors as well, maybe not.
Update on P8 Parkinson’s Disease.
@Gowtham_Rao what are next steps for this Phenotype?
Here’s what I think seem to be current status and next steps on this roadmap:
- Two proposed flavors of phenotypes: unanimity and tiered consensus.
- last week, we created two initial unanimity phenotypes (without and with meds)
- more about tiered consensus below
- Unaminity - proof of concept run results:
- inital results posted above by @Gowtham_Rao
https://data.ohdsi.org/PhenotypePhebruary2023_P8_ParkinsonsDisease/ - I have not seen the Atlas attrition tables for 309 and 310
- review of Cohort Diagnostics suggests the following:
- need to add concept Progressive Supranuclear Ophthalmoplegia to the “parkinsonism (neurodegenerative, nonPD) conditions” concept sets. I have updated in: Atlas-demo concept set 1872601 (2/13/23); not yet updated in atlas-phenotype.ohdsi.org yet
- more details about how phenotype behaves in two datasets require attention: jmdc and truven_mdcd in that phenotype does not result in expected male, older predominance demographics; it seems there is more secondary parkinsonism in both datasets overall, but still have to figure out why. From phenotype point of view, would like to identify if I am missing possible secondary parkinsonisms concepts that should be excluded in last 3 years; one way to approach this is to partially address is to create a unanimity cohort that will exclude ALL secondary parkinsonisms for all time (not just 3 years) and see if that results in expected demographics in these datasets. Another option worth exploring is if we should include a limitation in visit types to office visits (as the original phenotype paper did).
- Would benefit from guidance on next steps to consider how to address above issues and to consider additional analysis or cohorts to help refine this phenotype.
For tiered consensus criteria
- I agree that specialty seems to be challenging for P8 for now.
- Movement disorder specialty is not practical (once we have our own OMOP-CDM instance, we can post how much difference this might make)
- there remains value in including neurologist visits if possible. Again, in published algorithms for detecting PD with specificity, the tired consensus perfoms with higher sensitivity, specificity and PPV compared to unanimity algorthm.
- I believe there is still value in assessing a tiered consensus criteria
- even without a visit specialty criterion, a tiered criteria will allow a comparison between a full 3 year exclusion (unanimity) vs a tiered non-specialty consensus criteria.
There are likely 3 approaches to take to tiered consensus criteria: - (1) validate/review the 6 atlas-demo cohort defintions as proposed by @allanwu on 2/8/23 and run those
- (2) consider adaptation of those 6 atlas-demo cohort defintions and remove the specialty criteria and just focus on effect of the tiers year-by-year. suggested options for these 6 cohorts: “tiered consensus wo meds” (current), “tiered consensus w meds” “tiered non-specialty consensus wo meds” “tiered non-specialty consensus w meds” (only the first is available in atlas-demo currently).
- (3) I remain unsure how scalable the Phea approach is for networks tudy
@fabkury I reviewed the Phea package and I think I see what you are trying to do. At each time point (essentially visit), you identify if that particular visit is included in the cohort or not. I would add that if a particular visit is NOT included in the cohrt, then at that point, the person would be consided to drop out of the cohort. This raises very interesting possiblities in tracking the evolution of patients.
However, for the basic P8 implementation of the tiered consensus criteria (understanding you did not implement specialty yet), one would merely need to apply the SQL/Phea approach to the entry point (most recent) visit, look back 3 years, and either include or not include that person into the cohort or not.
And I remain interested in any of above cohorts, in the future, how we would be able to do an inference of incidence (earliest entry point). I didn’t quite follow @Patrick_Ryan post on this.