@allanwu Thank you so much for your leadership in driving this conversation, for your courage to contribute, and for the extremely clear and informative clinical description and current landscape assessment. I learned a lot by reading this. There’s clearly a lot of subtlely here with differential diagnoses and non-specific symptoms that make it a fun phenotyping challenge. Tagging @aostropolets and @callahantiff because we were just talking yesterdy about ideas of how to we leverage external knowledge on symptoms and differential diagnoses as part of the phenotype process, and this is a tremendous examplar to allow us to sink our teeth into.
Thank you also for providing a very clear rubric to follow to consider Possible / Probable / Definite cases of PD. It was very easy for me to follow your instructions that create a standard ATLAS cohort definition to do what you are looking for.
I’ll use this thread to illustrate the cohort definition design, because it provided a few opportunities to bust out some less-commonly-used tricks that others may enjoy learning about:
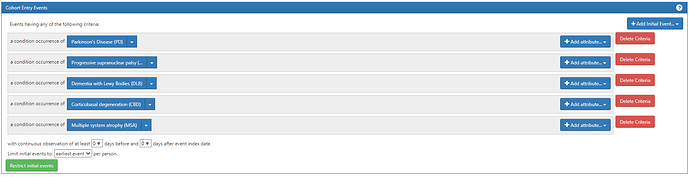
We start with the entry event, which is any condition occurrence of the Parkinsonism conditions (Parkinson’s disease, Progressive supranuclear palsy, Dementia with Lewy Bodies, Coritcobasal degeneration, and multiple system atrophy):
You’ll note that I created a separate conceptset for each of the Parkinsonism subtypes. That way we can reuse them as individual components, rather than having them all combined into one composite conceptset.
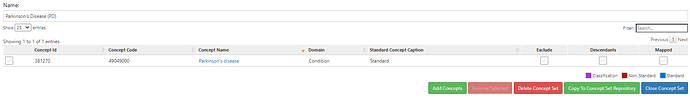
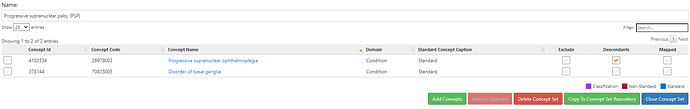
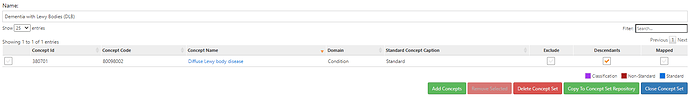
Those conceptsets were pretty straightforward to create based on the ICD codes that @allanwu provided. I’ll show the conceptset expressions just to highlight how concise they are (even though there can be many source codes that roll up to standards):
and for completeness, here’s the Parkinson’s drug conceptset:
and the ‘Parkinsonism confounder conditions’ conceptset we’ll use later
Ok, so now let’s walk through the inclusion criteria.
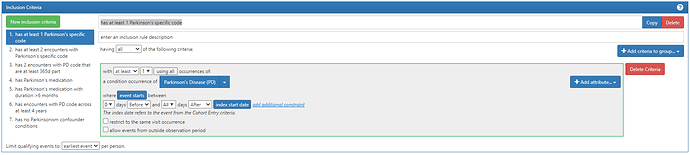
#1. has at least 1 Parkinson’s specific code:
Note above, we are able to ‘re-use’ that Parkinson’s conceptset that was in the entry event and now is used again here in this criteria. Since the entry was the earliest event of any of the Parkinsonism codes, we know that PD-specific code must be on or anytime after the index date.
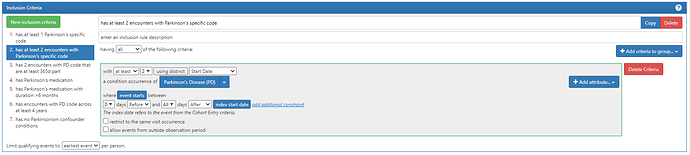
#2. has at least 2 encounters with Parkinson’s specific code
Note here, to requiring two separate dates with PD codes, changed the first componet to be ‘with at least 2’ (changed from 1) then clicked the button that said ‘using all’ and changed it to ‘using distinct’, then selected ‘Start Date’ from the dropdown.
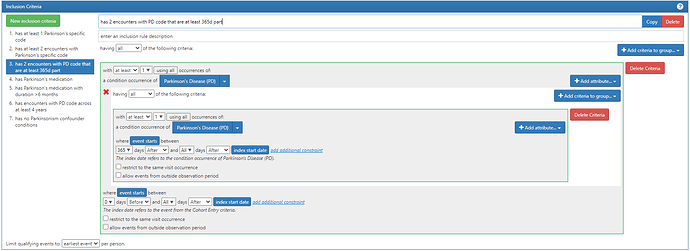
#3. has 2 encounters with PD code that are at least 365d part
Note here, I use the trick of a ‘nested criteria’ (which you find by clicking the ‘Add attribute…’ button and selecting the last item in the list. This allows us to ‘reset’ the index date for the criteria, and this case it means, we say, ‘must have at least 1 PD code which itself has at least 1 PD code that falls 365d after the first PD code’. This logic requires 2 codes with at least 365d gap between them.
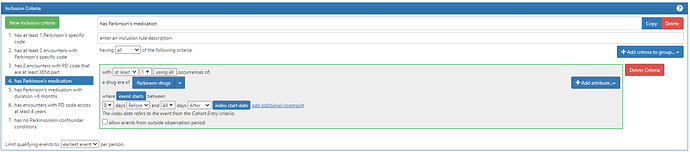
#4. has Parkinson’s medication
Two notes from above: 1) I have chosen to use ‘Drug Era’ but I could have used ‘Drug Exposure’ as the domain (you’ll see what I used Era in the next criteria). 2) I’m looking for drug after the entry event. It is possible that a person may have exposure PRIOR to their first observed diagnosis, particularly if we have prevalent cases of PD in our dataset. However, given Allan’s description and insight that drug may be used to ‘test for’ disease before diagnosis, I thought it better to focus on post-index exposures here.
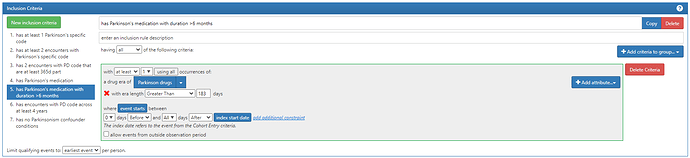
#5. has Parkinson’s medication with duration >6 months
Note this is the same criteria as #4, except that I added an extra attribute: ‘Add Era Length Criteria’, and set this to be ‘Greater Than’ 183 days. This is why I used the DRUG_ERA table, to take advantage of the pre-processing we do in the OMOP CDM to create episodes of continuous exposure. If we didn’t have that already in place, implementing this seemingly-simple logic ‘duration > 6 months’ would be extremely painful.
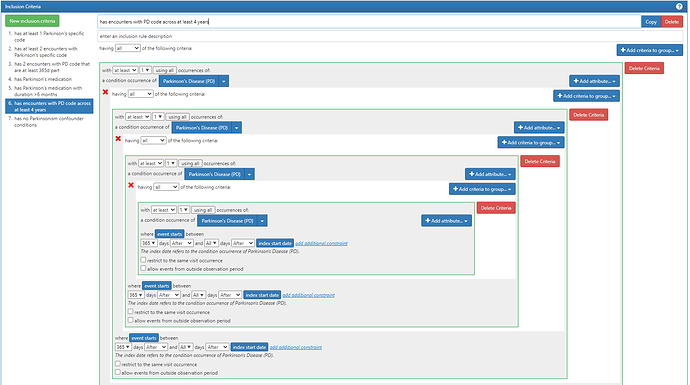
#6. has encounters with PD code across at least 4 years
This is probably the most complicated of the inclusion criteria. You’ll see I use the ‘nested criteria’ again, but I nest the nesting criteria 3 times. Basically, I’m saying: ‘you must have a PD code for which at least 365d later you have another PD code, for which >365d later there’s another PD code, for which >365d later, there’s another PD’. This guarantees that there’s at least 4 years with a PD code appearing.
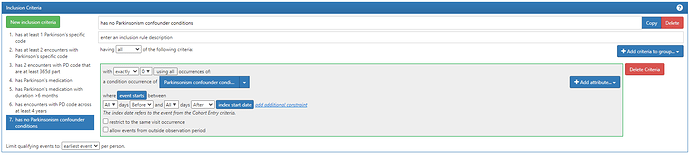
#7. has no Parkinsonism confounder conditions
Note, we look for the absence of any confounder conditions and we use all time pre- and post-index.
There were two criteria that @allanwu proposed that I did not implement, and I’ll discuss both briefly:
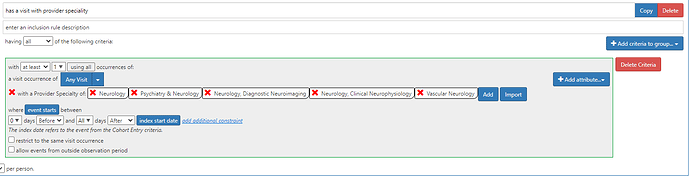
“(7) with at least one code coming from neurologist (neurology / movement disorders visit)” - if we want to implement this, we need to define how to recognize neurologist participation in care. On a admittedly quick scan, I couldn’t find any particular CPT/ICD10PCS codes that specifically highlight procedures that ensure its a neurologist involvement (even though there are many codes that involve neurological evaluation). As a community, we are still harmonizing on provider specialty, and I know many databases either do not provide this information or if its particularly noisy (including not mapped into visit occurence table). Many of the NUCC concepts aren’t specific to Neurology (they combine Psychiatry and Neurology), so I’m just out of my depth to implement this reliably. (Just as a demonstration, one can use ATLAS to require visit with Provider Specialty like this:)
“(8) + ratio of PD to other Parkinsonism confounder code is >2:1” - this is not a function currently supported in ATLAS. But, personal opinion, I find these types of heuristics hard to defend on any sort of clinical grounds, usually they are serving as some crude proxy for some other idea that one has in mind that may be able to modeled more appropriately. In this particular case, we also look for ‘no parkinsonism confounder codes’ and we see that doesn’t impact the cohort substantially (see below) so I don’t think we need to try to get too fancy with this rule.
And, here’s the results from the MarketScan CCAE database. I’m showing here the attrition table from ATLAS (go to Generation tab, Generate the cohort on your database, then click ‘View Reports’ button, you can then toggle between ‘Intersect view’ and ‘attrition view’). Intersect view helps understand the independent impact of each criteria. Attrition view lets you see the inclusion criteria applied in sequential order. Given the framework Allan provided, I’ll show the attrition view here:
We can see that we start with 105,243 patients with a Parkinsonism code, of which 83.17% (87,532) have at least one Parkinson’s specific code. Of those, 63k have at least 2 encounters with a PD code, and 33,757 have 2 codes that are more than 365d apart. 87% (29k/33k) of these patients had at least one Parkinson’s disease medication, and 84% (25k/29k) of those with a drug had at least one duration of exposure greater than 6 mo. The criteria had the largest proportional impact on the cohort was requiring at least 4 encounters across 4 years: only 9,238 met this criteria. Note, this can be partly a reflection of the database, which contains privately insured patients, who there is both an issue with persons not having long continuous observation and also an issue with persons > 65 transitioning to Medicare. The last criteria “no Parkinsonism confounder conditions” did not have a major impact, with >91% of those patients remaining for the final cohort count of 8,480.
Note, now that we have this one cohort definition that implements almost all of the Wu criteria (modulo Neurologist visit and majority rules confounders), it is easy to create a definition that relaxes the criteria to find all ‘Probable’ or ‘Possible’ cases. You just have to delete the criteria that are not relevant. For illustration purposes, I’ve provided 3 cohorts on ATLAS-phenotype, and I’ll ask @Gowtham_Rao if he could be so kind as to run CohortDiagnostics on these 3 definitions, so that we can see how the patient characteristics may vary across these variants:
-
[Phenotype Phebruary][PD] Persons with Parkinson’s disease - Wu definite
-
[Phenotype Phebruary][PD] Persons with Parkinson’s disease - Wu probable
-
[Phenotype Phebruary][PD] Persons with Parkinson’s disease - Wu possible
I’d be really curious to see if PheValuator could give us insights about the sensitivity/specificity tradeoffs between the definite/probable/possible classification that @allanwu has proposed. @jswerdel this may be a fun demonstration case where the cohorts are more akin to PheValuator 1.0 framing of ‘prevalent chronic disease’ but for which PheValuator 2.0 should still be applicable (just with using longer feature windows that extend beyond the acute post-visit window)
Fun stuff! I’m eager to hear from others about where we go from here…