Dear community, I am happy to share the R package I created for electronic phenotyping, called Phea. The name losely stands for “phenotyping algebra.” It provides a framework for phenotyping that:
- Is based on formulas, for example
bmi = weight / (height * height), orplatelet_drop = platelet_current - platelet_previous - does not use SQL joins to combine the data elements needed to calculate the formula.
Phea is on GitHub at https://github.com/fabkury/phea/:
devtools::install_github('fabkury/phea')
library(phea)
Under the hood, Phea is just a SQL query builder. All computation is done inside the SQL server, or you can ask to get just the final SQL code of the query. For those in the know, Phea leverages the dbplyr/dplyr lazy table infrastructure in the R language, as well as inherits the principles of tidy data that underpin them.
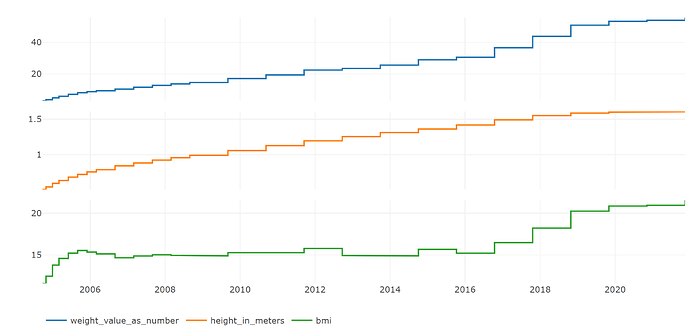
Phea shines for efficiently computing clinical scores such as a disseminated intravascular coagulation score, or the ASCVD Risk Estimator+ (to name just two), for any number of patients at all points in time. The results for one patient look like this:
Other use cases are also possible, and in fact, the formulas are allowed to contain any SQL expression that is valid inside a SELECT statement. For example, CASE WHEN ... constructs, or any function call supported by the server. Phea was built with OMOP CDM in mind, but is not restricted to it.
-
For a very brief look at how Phea can be used, please see the vignette computing body mass index.
-
For an explanation of the intuition behind Phea, and a look at its features, see getting started with Phea.
While Phea is a standalone tool, if you need to use its results inside Atlas, one approach is to use it to produce novel records, then ETL those back into the dataset using custom concept IDs.
Besides that, there are a few other perspectives on Phea that I look forward to presenting to its potential users. I will be happy to post here in the forum, later on, about:
- Using the result of prior formulas inside other formulas.
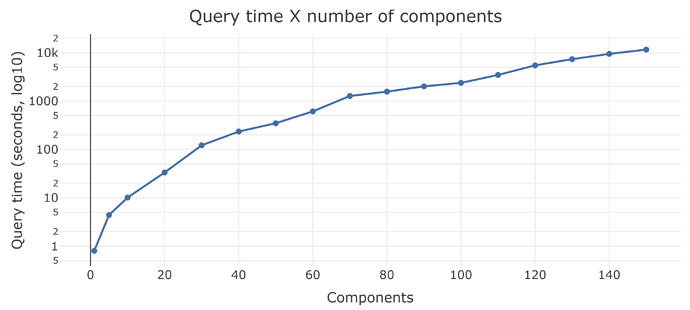
- Stress-testing Phea with formulas containing hundreds of components, each coming from a different SQL query; as well as hundreds of consecutive layers of formula->result->another formula.
- Computing formulas with multiple different records from the same SQL query, e.g. a patient’s second instance of a diagnosis, or the hemoglobin A1c level from 60 days ago alongside its value today.
- Computing formulas with Boolean components, as opposed to numeric ones.
- Phenotyping entities other than patients, such as providers or care sites.
I am more than happy to receive any comments, here or via email.
Kindest regards, and happy Black Friday for those here in the US.