My R package Phea can help compute changes in lab values over time, for example “increase in serum creatinine by >= 0.3 mg/dL within 48 hours”, or “ratio of >= 1.5 within 7 days”.
I wrote a vignette to walk you through how to do that using the package. Please see it here.
This is how the phenotype looks like:
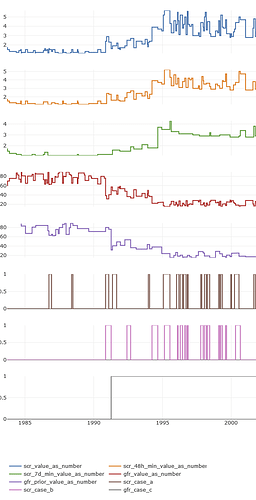
And here is how the plot of the scr_change phenotype looks like for a single patient:
My vignette is limited to data that Synthea™ can produce. I hope the demonstration is clear enough so that anyone can easily update, for example, the concept IDs, or how units of measurement are identified. If anyone would like to talk to me about this over email or a call, just let me know!
Phea’s phenotypes are just SQL queries. The final SQL code for scr_change is in the vignette. You can get that query and take it elsewhere, if wanted.