Team:
Day 5 of Phenotype Phebruary. Still lots of methodological topics to discuss and disease areas to investigate. Today, I’ll try to start a conversation of the phenotype that was most highly voted on across our community: Alzheimer’s disease.
Clinical description:
Alzheimer’s disease is a progressive neurodegenerative disorder and the most common cause of dementia (loss of cognitive functions interfering with daily activities), representing 60-80% of cases (according to Alzheimer’s Association). Intitial symptoms of Alzheimer’s disease may be short-term memory loss and other difficulties associated with mild cognitive impairment, such as word-finding, visual/spatial issues, and general confusion. Diagnosis of Alzheimer’s disease may involve neurological exam, including brain MRI or CT scans, to identify other potential causes of dementia other than Alzheimer’s, and mental cognitive status tests. Drugs approved for use in Alzheimer’s disease include cholinesterase inhibitors (such as donepezil, galantamine, or rivastigmine) and memantine, which are primarily aimed at treating cognitive symptoms. In 2021, aducanumab was approved by US FDA on the basis of clinical trial data suggesting reduction of amyloid beta plaque. Alzheimer’s disease risk increases with age, with most cases detected after 65 years old. Prevalence of AD is higher in females than males, though that is attenuated by female longer life span. It is one of leading causes of death globally, and second-leading cause in high-income countries (WHO).
Phenotype development:
I’ve mentioned in prior posts that a valuable starting point for phenotype development can be the published literature, and I’ve shown how you - provided that a journal article supplied enough details - you can replicate their algorithms using OHDSI tools. But I want to take a digression here for a little rant: if observational researchers all need to develop phenotypes to conduct our analyses and should all review prior literature as part of our research process, then why is so hard to search for publications of observational research and extract out the phenotypes that were previously used? If phenotypes are so central to the integrity of our research, then as a research community, why do we accept short freetext descriptions of phenotypes in manuscripts, sometimes without list of codes and often without a complete specification of the logic that was used to implement them? And for those of us promoting increased transparency, when we try to add additional detail in supplemental materials, why do we often format it in ways that make it painful for others to re-use without extensive manual curation? When I read a paper that I’m excited by and want to replicate, what I wouldn’t give to just get a JSON specification of conceptsets and cohort logic, not only to save me effort but also to prevent error and avoid ambiguity.
The first step is actually finding papers that contain phenotypes to consider. Now, PubMed is an AMAZING resource; my wife is an English professor, so I get to see what research can require if one doesn’t have a centralized repository of published scholarship (it looks aweful), and I never take for granted the ability to quickly identify and retrieve scientific references. Thank you NLM! But, perhaps because we are in a fairly niche space, the maturity of observational database research publications and their content isn’t always straightforward to capture.
The search problem here is: Find all observational database studies that contain a phenotype algorithm for Alzheimer’s disease. A plea to our community: if anyone has a great PubMed search strategy for this task, please share it by replying to this post.
I’ll share the search strategy I use, which is something that I started working on back in the OMOP days and cleaned up and improved with the help of Gayle Murray, our wonderful library expert at Janssen.
(“Alzheimer disease”[MeSH Terms] OR “Alzheimer”[Title/Abstract])
AND
((“retrospective cohort”) OR (Epidemiology[MeSH Terms]) OR (Epidemiologic Methods[MeSH Terms]) OR (phenotype[Title/Abstract]) OR (insurance) OR (claims) OR (database) OR (Diseases Category/epidemiology[MeSH Terms]) OR (Validation Study[Publication Type]) OR (Validation Studies as Topic[MeSH Terms]) OR (Sensitivity and Specificity[MeSH Terms]) OR (Predictive Value of Tests[MeSH Terms]) OR (Reproducibility of Results[MeSH Terms]) )
AND
((Medicaid) OR (Medicare) OR (Truven) OR (Optum) OR (Medstat) OR (“Nationwide Inpatient Sample”) OR (“National Inpatient Sample”) OR (PharMetrics) OR (PHARMO) OR (ICD-9[Title/Abstract]) OR (ICD-10[Title/Abstract]) OR (IMS[Title/Abstract]) OR (“electronic medical records”[Text Word]) OR (Denmark/epidemiology[MeSH Terms]) OR (Veterans Affairs[Title/Abstract]) OR (“Premier database”[Title/Abstract]) OR (“National Health Insurance Research Database”[Title/Abstract]) OR (Outcome Assessment[Title/Abstract]) OR (“insurance database”[Title/Abstract]) OR (Database Management System[MeSH Terms]) OR (Medical Records Systems, Computerized[MeSH Terms]) OR (“Positive predictive value”[Title/Abstract]) )
NOT
(“Clinical Trial”[pt] OR “Editorial”[pt] OR “Letter”[pt] OR “Randomized Controlled Trial”[pt] OR “Clinical Trial, Phase I”[pt] OR “Clinical Trial, Phase II”[pt] OR “Clinical Trial, Phase III”[pt] OR “Clinical Trial, Phase IV”[pt] OR “Comment”[pt] OR “Controlled Clinical Trial”[pt] OR “Letter”[pt] OR “Case Reports”[pt] OR “Clinical Trials as Topic”[Mesh] OR “double-blind”[All] OR “placebo-controlled”[All] OR “pilot study”[All] OR “pilot projects”[Mesh] OR “Prospective Studies”[Mesh] OR “Genetics”[Mesh] OR (“Genotype”[Mesh]) OR (biomarker[Title/Abstract]))
Basically, the search string is a combination of 1. the phenotype target (varients in MeSH terms or freetext), 2. a list of markers to indicate the type of study, 3. a list of terms to verify that its a database study, and 4. exclusions to remove non-observational research.
This returns back 1,009 results, most of which are not helpful for that I’m trying to accomplish. So, I’ve got a sensitivity/specificity problem with my search string…just like my phenotypes ![]()
The second step is to extract phenotype algorithms from the relevant papers. Here are a few papers that were near the top of the list or were , which are useful for this discussion:
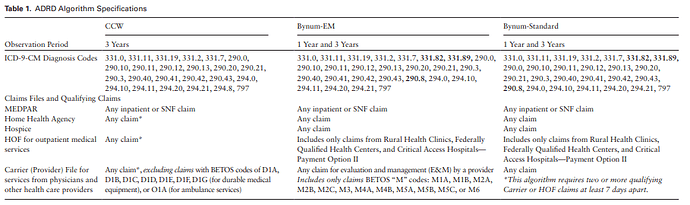
They provide a brief description of their three algorithms with the following table to enumerate code choices (shown below) . In their decription, they state that they are starting with a codeset from prior literature, but then they evaluate modifications by “adding diagnosis codes for dementia with Lewy Bodies (331.82), other cerebral degeneration (331.89), and other nonspecified senile psychosis (290.8) based on discussion with experts in the field”
The authors describe that “To ensure the correct list of codes, we began with a list of ADRD
diagnostic codes used in the literature”, but then, “We also considered codes for other medical
conditions, such as other degenerative conditions, delirium”.
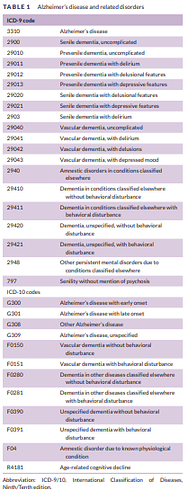
The authors describe " AD tends to be under‐coded in claims data and thus we used the broader definition of “dementia” as our primary endpoint, which includes AD as well as generic conditions of “senility,” “degenerative brain disorder,” and others, but excludes Lewy body dementia, drug‐ and alcohol‐induced dementia, or dementia caused by concussions or syphilis, among others. A full list of the included concepts and mapped ICD‐9‐CM and ICD‐10‐CM codes can be found in Appendix Table.", and then provide this:
| Code | Name | Vocabulary |
|---|---|---|
| Alzheimer’s Disease codes | ||
| 331.0 | Alzheimer’s disease | ICD9CM |
| G30.0 | Alzheimer’s disease with early onset | ICD10CM |
| G30.1 | Alzheimer’s disease with late onset | ICD10CM |
| G30.8 | Other Alzheimer’s disease | ICD10CM |
| G30.9 | Alzheimer’s disease, unspecified | ICD10CM |
| Dementia Codes | ||
| 290.0 | Senile dementia, uncomplicated | ICD9CM |
| 290.10 | Presenile dementia, uncomplicated | ICD9CM |
| 290.11 | Presenile dementia with delirium | ICD9CM |
| 290.12 | Presenile dementia with delusional features | ICD9CM |
| 290.13 | Presenile dementia with depressive features | ICD9CM |
| 290.20 | Senile dementia with delusional features | ICD9CM |
| 290.21 | Senile dementia with depressive features | ICD9CM |
| 290.3 | Senile dementia with delirium | ICD9CM |
| 290.40 | Vascular dementia, uncomplicated | ICD9CM |
| 290.41 | Vascular dementia, with delirium | ICD9CM |
| 290.42 | Vascular dementia, with delusions | ICD9CM |
| 290.43 | Vascular dementia, with depressed mood | ICD9CM |
| 294.0 | Amnestic disorder in conditions classified elsewhere | ICD9CM |
| 294.10 | Dementia in conditions classified elsewhere without behavioral disturbance | ICD9CM |
| 294.11 | Dementia in conditions classified elsewhere with behavioral disturbance | ICD9CM |
| 294.20 | Dementia, unspecified, without behavioral disturbance | ICD9CM |
| 294.21 | Dementia, unspecified, with behavioral disturbance | ICD9CM |
| 331.0 | Alzheimer’s disease | ICD9CM |
| 331.11 | Pick’s disease | ICD9CM |
| 331.19 | Other frontotemporal dementia | ICD9CM |
| 331.2 | Senile degeneration of brain | ICD9CM |
| 331.9 | Cerebral degeneration, unspecified | ICD9CM |
| 797 | Senility without mention of psychosis | ICD9CM |
| F01.50 | Vascular dementia without behavioral disturbance | ICD10CM |
| F01.51 | Vascular dementia with behavioral disturbance | ICD10CM |
| F02.80 | Dementia in other diseases classified elsewhere without behavioral disturbance | ICD10CM |
| F02.81 | Dementia in other diseases classified elsewhere with behavioral disturbance | ICD10CM |
| F03.90 | Unspecified dementia without behavioral disturbance | ICD10CM |
| F03.91 | Unspecified dementia with behavioral disturbance | ICD10CM |
| G30.0 | Alzheimer’s disease with early onset | ICD10CM |
| G30.1 | Alzheimer’s disease with late onset | ICD10CM |
| G30.8 | Other Alzheimer’s disease | ICD10CM |
| G30.9 | Alzheimer’s disease, unspecified | ICD10CM |
| G31.01 | Pick’s disease | ICD10CM |
| G31.09 | Other frontotemporal dementia | ICD10CM |
| G31.1 | Senile degeneration of brain, not elsewhere classified | ICD10CM |
| G31.89 | Other specified degenerative diseases of nervous system | ICD10CM |
| G91.4 | Hydrocephalus in diseases classified elsewhere | ICD10CM |
[Related aside, but this is a very nice paper from collaborators within our OHDSI community, @Dave_Kern and @scepeda. It shows how OHDSI tools and best practices can be used to conduct a population-level effect estimation study across two claims databases to test a biological hypothesis. Nice examples of large-scale propensity score adjustment to achieve covariate balance and use of negative controls to evaluate residual confounding]
So, three papers trying to use claims data to study Alzheimer’s disease and related dementias, three different formats for how the codelists are shared, and indeed, three different lists of codes. But how does one go about finding the subtle differences between the codelists? And how does one determine if the differences are actually impactful?
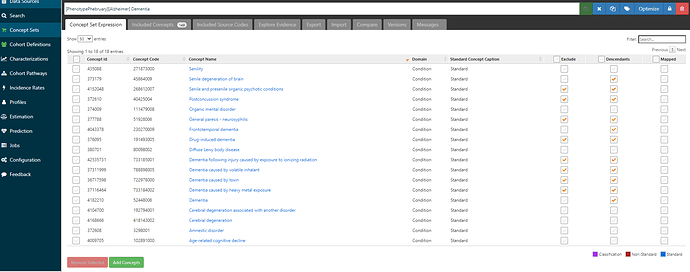
One approach that I’ve created as a bit of a habit for myself, when reviewing the prior literature and trying to create a conceptset: I think about the literature as the union of all codes that I may be interested in, and I make a conceptset that maps from the source codes provided to their corresponding OHDSI vocabulary standard concepts, so that ultimately I end up with a conceptset expression that subsumes all the codes from the literature (and by virtue of the SNOMED hierarchy and source code mappings, may also sweep in a few extra codes that the literature had missed).
What does my standardized conceptset expression look like here, to capture all the ideas from McCarthy, Jain and Kern?
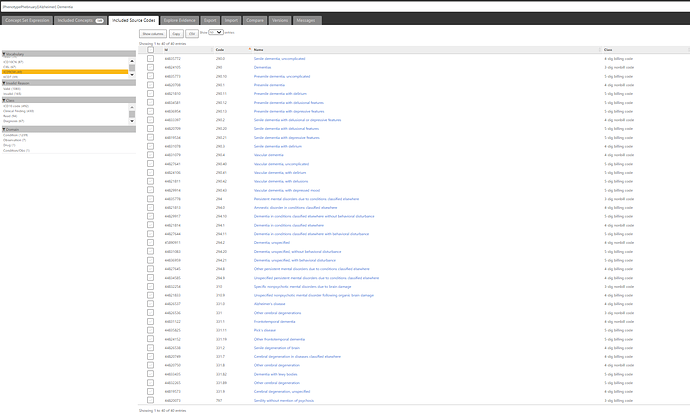
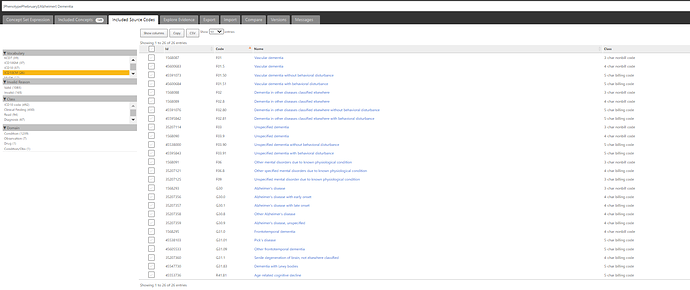
If we look at the ICD9CM codelist covered, we can see 40 codes provided:
And for ICD10CM, we’ve got 26 codes:
But, you don’t want these teeny tiny screenshots any more than I want to have to manually type out code-by-code from the various forms in the publication. What you want is something that is both human-readable and computer-executable. Here’s the JSON specification for the dementia conceptset (xml filetype only to enable it to be uploaded here on the forums)
Phenotype Phebruary dementia conceptset.xml (9.6 KB)
Once I have created my conceptset covering the literature, then I can continue with some of the other steps that we’ve discussed earlier this week, like using PHOEBE to see if any other concepts are recommended, creating cohorts using those conceptsets, running CohortDiagnostics or PheValuator to evaluate the cohorts, etc.
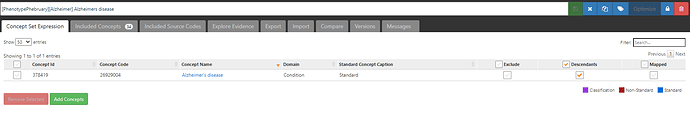
Now, the ‘dementia’ conceptset may look not-so-straightforward, and that’s in part because the various papers disagreed on assorted edge cases of other types of dementias and cognitive decline. But the conceptset for ‘Alzheimer’s disease’ is a thing of beauty:
One concept, plus its descendants, covers 34 standard concepts and fully subsumes all the ICD9CM and ICD10CM codes highlighted by the papers.
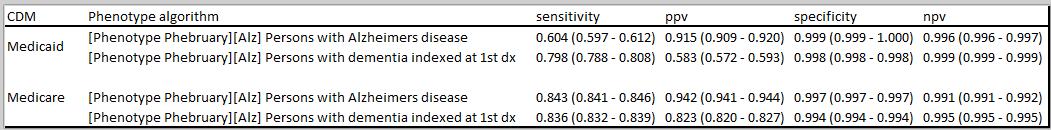
I want to go back to the paper by @Dave_Kern and @scepeda . They do something quite smart in their analysis: they are interested in the target of Alzheimer’s disease, but they noted from prior literature that AD algorithms had low sensitivity but good specificity. But we also know from prior literature that Alzheimer’s makes up 60-80% of dementia cases. So, one can consider a dementia phenotype to be a ‘more sensitive, less specific’ phenotype algorithm for Alzheimers. Given that these two alternative algorithms represent a logical tradeoff in measurements errors, they executed their analyses using both definitions. And when they generated consistent estimates with both approaches, they could more confidently conclude that their finding wasn’t just a bias due to one algorithm. In situations where there is no one clear right algorithm, and the impact of alternative definitions can be substantial (as was here), then this idea of ‘bounding’ your analyses with a sensitive algorithm and a specific algorithm seems like a good strategy to consider when trying to manage worries of measurement error.
Resources built today to help this discussion (conceptsets are embedded within these cohorts):
- ATLAS-phenotype link for definition of ‘Persons with Alzheimers disease indexed at first diagnosis’
- ATLAS-phenotype link for definition of ‘Persons with Alzheimers disease’, indexed on first diagnosis that has a follow-up diagnosis within 365d
- ATLAS-phenotype link for definition of ‘Persons with dementia indexed at first diagnosis’
- ATLAS-phenotype link for definition of 'Persons with dementia, indexed on first diagnosis that has a follow-up diagnosis within 365d
I know several of you have done research in Alzheimers and dementia. Share your phenotype experience. What did you learn? How can future work be improved? Is there anything that I’ve said here that resonated or that you disagree with? Let’s discuss!