Today, I’d like to a little side step from the problem of creating phenotypes de novo, and talk about how to try to implement a phenotype algorithm using OHDSI tools based on an existing description from some external material, just as a publication. I’ll use the phenotype of Atrial Fibrillation to demonstrate some tips and tricks, but I hope you’ll see that the steps I’ll follow here are completely transportable to whatever phenotype you may want to be working on.
Clinical description:
Atrial fibrillation (AFib) is an abnormal heart rhythm, often with irregular beats in the atrial chamber. CDC has a nice animated gif to illustrate what this means: https://www.cdc.gov/heartdisease/atrial_fibrillation.htm. Initial symptoms of AFib may include feeling of irregular heartbeat or palpitations, lightheadedness, fatigue, dyspnea, and chest pain. AFib is frequently comorbid with other cardiovascular conditions, such as hypertension, coronary artery disease, and pericarditis. Patients with AFib are at higher risk of cardiovascular complications, including heart failure and ischemic stroke. Afib is most commonly diagnosed with electrocardiogram, though echocardiogram can also be used to evaluate Afib and valvular defects. AFib can be classified as ‘first detected’, ‘paroxysmal’, ‘persistent’, ‘longstanding persistent’, and ‘permanent’ based on the frequency and duration of abnormal heartbeat episodes. Common pharmacologic treatment may include beta blockers, calcium channel blockers, amiodarone, and anticoagulants (such as warfarin, heparin, or a direct oral anticoagulant (DOAC) such as apixaban, rivaroxaban, and dabigatran). Other treatments can include electrical cardioversion and catheter or surgical ablation. AFib is more common in men than women, and the risk of AFib increases with age.
** Phenotype development: **
AFib is a common target for investigation in observational databases, both as a disease that impacts future outcomes and also as an indication for various treatments that are regularly evaluated for comparative effectiveness and safety.
As a case in point, just PubMed ““atrial fibrillation” AND (“claims” OR “electronic health records”)” and you’ll see >1,000 articles! You’ll find many recent observational studies in high-profile journals. Articles like Ray et al, " Association of Rivaroxaban vs Apixaban With Major Ischemic or Hemorrhagic Events in Patients With Atrial Fibrillation", in JAMA in Dec2021 (which seems like a nice target for an OHDSI replication, wink wink @agolozar ). Or Kim et al, " Machine Learning Methodologies for Prediction of Rhythm-Control Strategy in Patients Diagnosed With Atrial Fibrillation: Observational, Retrospective, Case-Control Study" in JMIR Med Inform Dec 2021 (which incidentally doesn’t say it used an OMOP CDM instance, but has a supplemental list of codes used that looks suspiciously like they extracted it out of an OHDSI vocabulary conceptset expression @mgkahn ). Another end of year paper that connects AFib with health equity, "Sex differences in treatment strategy and adverse outcomes among patients 75 and older with atrial fibrillation in the MarketScan database" by Subramanya et al in BMC Cardiovascular Disorders (wink @noemie ). Actually just this month, there’s a paper in J Am Heart Assoc, " Comparative Safety and Effectiveness of Sotalol Versus Dronedarone After Catheter Ablation for Atrial Fibrillation" by Wharton et al.
If we look at the Wharton paper, you’ll find a typical textual description of their phenotype: " Patients with a diagnosis of AF were identified within the study period between January 1, 2013 and March 31, 2018 using International Classification of Diseases , Ninth Revision and Tenth Revision (ICD‐9 and ICD‐10 ) codes." The authors provide a supplemental materials file that provides the specific list of ICD codes:
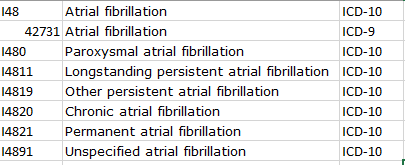
In the Subramanya paper, you get a phenotype description like this: “AF was diagnosed using International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes, 427.31 and 427.32 for one inpatient or two outpatient claims between 7 days and a year apart, in any position”
Note, both of these two papers are using a MarketScan claims database, and both are providing ICD9 codes, though they disagree both on the codelist (Wharton is not using ICD9 427.32 ‘Atrial flutter’) and logic (Subramanya requiring one inpatent or two outpatient diagnoses, whereas Wharton requires only one diagnosis in any setting)… If only we had some tools that could us empriically evaluate the consequences of these phenotype algorithm differences ![]()
Regardless of whether one is ‘better’ than the other, both have provided a definition and a codelist, so my task for today is to show how we could use OHDSI tools to replicate the author’s intent.
Let’s start with Wharton et al:
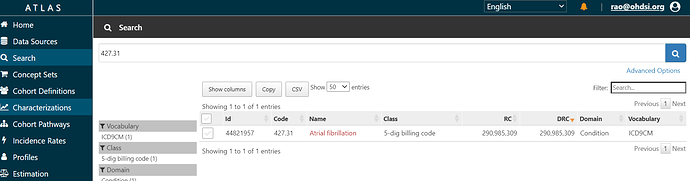
We can start with the codelist provided, and construct a conceptset expression. In ATLAS, you can search the vocabulary for any string, whether it be a term like ‘atrial fibrillation’ or a source code like ‘427.31’ (or a concept_id if you happened to have it handy). Here, I’ll assume all I’ve got is what Wharton has given us in their supplemental materials, and I’ll start to search for the 1 ICD-9 code, 427.31.
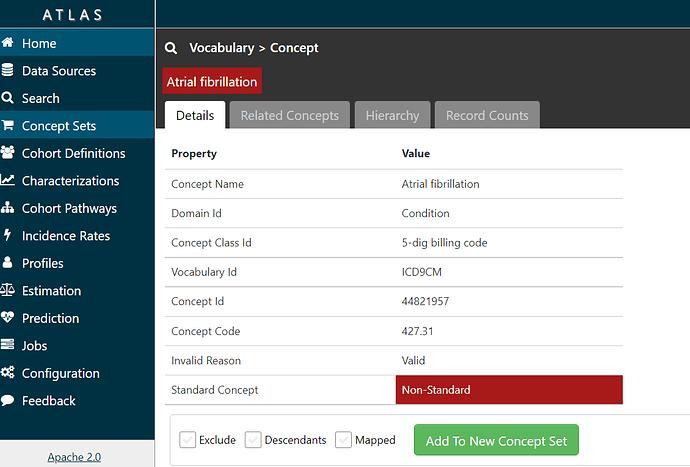
If I click on the name ‘Atrial Fibrillation’, I can see information about the concept from the OHDSI vocabulary:
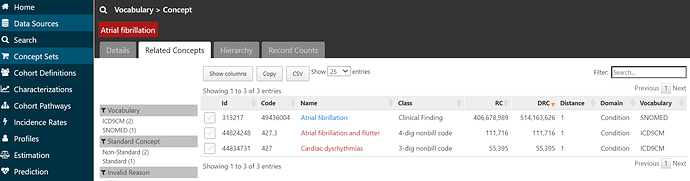
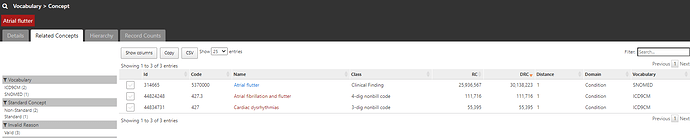
and if I click on the ‘Related Concepts’ tab, I can see that the code 427.31 has three relationships, one to the SNOMED standard concept called ‘Atrial fibrillation’ (ID = 313217).
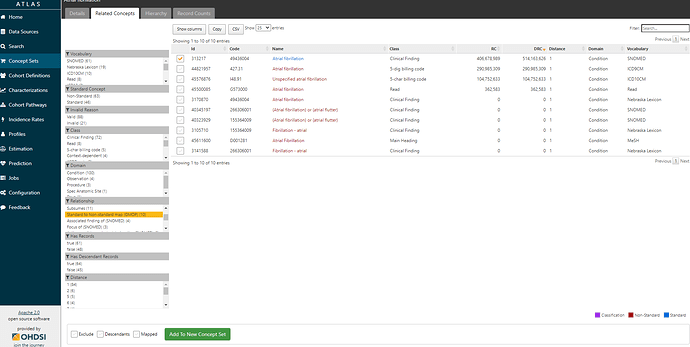
If I click on that standard concept 313217 and look at its ‘Related Concepts’, I can see that now that I’m in the standards space, I have a much wider collection of relationships (109 in all, including 46 standard ancestral relations, and 63 non-standard concepts mapped). If I restrict to see source codes that are directly mapped to ‘Atrial fibrillation’ (by filtering to relationship = ‘Standard to Non-standard map (OMOP)’, I see there’s not only the one ICD9CM code I started with (427.31), but also now one ICD10CM code I48.91 ‘Unspecified atrial fibrillation’, and one READ code G5730000 ‘Atrial fibrillation’. Just by mapping from the ICD9 source code to a standard concept, I’ve learned of codes in two other source vocabularies that I can now look into. So, I’m going to start a conceptset using this standard concept, by clicking the checkbox next to the left of the concept and then pushing the green ‘Add To New Concept Set’ button below.
I can click on Concept Sets tab to the left to see where I’m at right now:
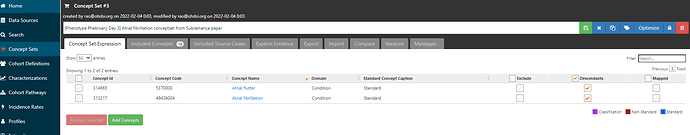
Now, I have a choice to make. I could repeat the same process I just did above for each of the source codes (e.g. Wharton et al provided seven ICD10 codes, all in the I48 range, noting that I48.91 is one of those codes and is already identified by our standard concept). The other approach is that I can use the power of the OHDSI vocabulary ancestry. If I expand my conceptset expression by checking the ‘Descendants’ box, then we can see that I have broadened out to 13 included concepts.
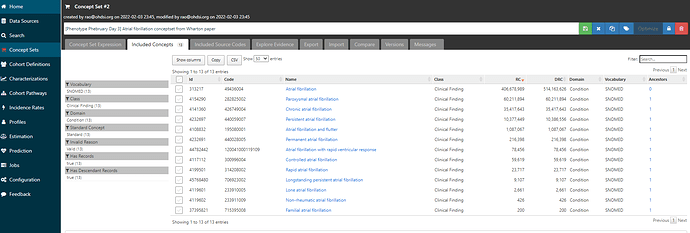
Clicking on the ‘Included concepts’ tab and sorting by the record count (RC), we can see that the concept ‘Atrial fibrillation’ is the most common concept observed, but there are more than 10 million records for ‘Paroxysmal atrial fibrillation’, ‘Chronic atrial fibrillation’, ‘Persistent atrial fibrillation’, all of which sound consistent with the clinical classification above as well as the terms that Wharton et al. had picked up from the ICD10 world.
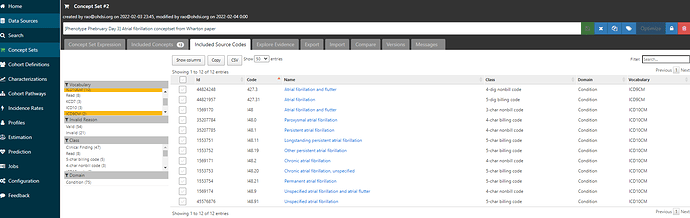
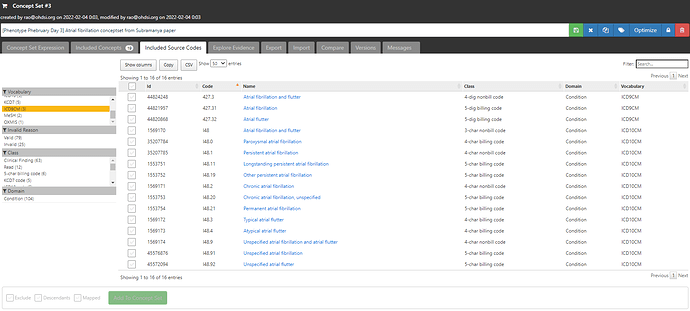
If I now move over to the ‘Included Source Codes’ tab and filter to only ICD9CM and ICD10CM, we can see that using only one standard concept, ‘Atrial fibrillation’ + descendants identifies for me 10 ICD10CM codes, all in that I48 series, and 2 ICD9CM codes (the 427.31 code I started with, and it’s parent 427.3 ‘Atrial fibrillation and flutter’):
But wait, Wharton only had 7 ICD10CM codes, so what’s the discrepancy? Well, Wharton didn’t list out all the 4-digit non-billing codes: they didn’t have I48.1 ‘Persistent atrial fibrillation’, but they had its descendant I48.11 ‘Longstanding persistent atrial fibrillation’; they didn’t have I48.2 ‘Chronic atrial fibrillation’, but they did include I48.20, ‘Chronic atrial fibrillation, unspecified’. Now, we could debate the merits of including non-bill codes and whether you want something in your list that may not be reimbursed, but I don’t think anyone could question that all of the ICD10 source codes listed above clearly belong clinically to the idea of ‘Atrial fibrillation’. And isn’t it pretty cool that we recovered the entire Wharton codeset by just using the OHDSI vocabulary mappings, and without having to search for each of the 8 codes one-by-one? All you need is this one clean conceptset: ATLAS
But what about Subramanya and their use of ICD9 427.32? This code is ‘Atrial flutter’, and hopefully to the disagreement to noone, maps to the standard SNOMED concept of ‘Atrial flutter’:
So, lets add that concept to our conceptset expression, and see it’s impact:
If we look at the included source codes from ICD9CM and ICD10CM now, we have 3 ICD9CM codes (both of the two codes mentioned, plus their parent 427.3, and we’ve also identified 3 additional ICD10CM codes, all in that same I48 series: I48.3, I48.4, I48.92:
Now, whether you want to include atrial flutter in your definition of atrial fibrillation, that’s a clinical choice, one which the material consequences can be empirically investigated using tools like CohortDiagnostics. But we can see how two papers starting with two different source code lists can be quickly implemented as OHDSI conceptset expressions, then those comparisons can be made using these standardized building blocks.
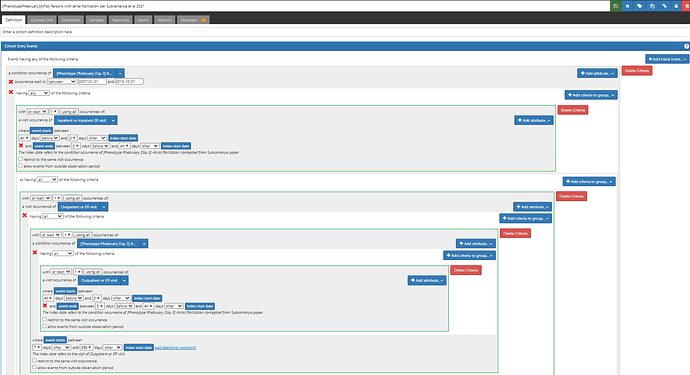
With these conceptsets in hand, we can go build the logic of the cohort. (note, for this demonstration, I’m only focusing on the AFib portion of the published studies, but the papers detail additional inclusion criteria for specifying their target populations for their questions of interest, I’m not covering that here).
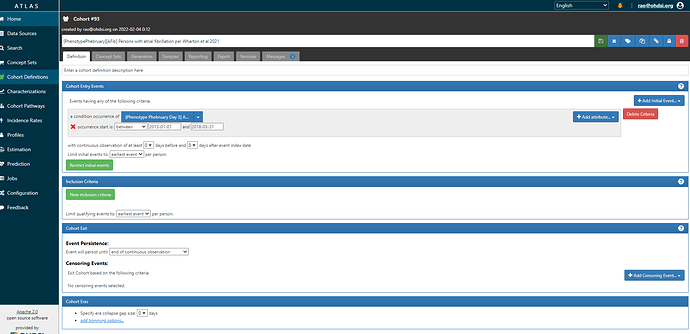
First, Wharton et al. It’s pretty simple because they just look for an occurrence of a code in their AFib list with a date between 1Jan2013 and 31Mar2018 . (ATLAS-phenotype link here)
Second, Subramanya et al, a little trickier, only because they want one inpatient or two outpatient codes that are 7d to 365d apart. The notion of ‘inpatient claims’ and ‘outpatient claims’ is very US claims oriented, and it can even be a little bit ambiguous for those less familiar in that emergency room visits may generate outpatient claims. So, when I see this type of ask, I try to genericize it to something that could be more broadly applicable across different data and geographies. Instead of claims, we could say ‘one AFib diagnosis occurring in a hospital admission’ OR ‘two Afib diagnoses that occur in an outpatient of ER visit, separated by 7d to 365d’. ATLAS-phenotype link here.
Usually, whenever I’m starting to consider phenotyping a new disease target that I haven’t phenotyped before, my first step is to look into PubMed to review prior publications, see how they developed their phenotype logic and what codes they used (and what, if any, validation information is available). Since most publications don’t (yet) use standard concepts from the OHDSI vocabularies, that means I often have to follow the source code → standard concept mapping, and then assess the impact of using a standard conceptset expression that subsumes the source codes that I started with. Because we have to do this mapping maneuver, people often get anxious that there’s going to be some information loss or noise introduced into the process, but @hripcsa has shown nicely in JAMIA 2018 that this mapping often has minimal impact vs .using source codes alone. My personal experience is that way more times than not, the mapping is either loss-less or actually identifies extra codes that I would want in my definition (like the extra ICD10 for Wharton above), so there’s a value add. But, each phenotype needs to be assessed on its own merit. Publications can be a good starting point for your own research, and the OHDSI tools can help you implement and evaluate your starting point, whether your aim is to exactly replicate the published code lists, or rather emulate their clinical intent (and try to improve upon their codeset or logic).