We have decided to submit the drug resistant epilepsy phenotype that was recently published in Epilepsia and presented at the OHDSI symposium for review by the phenotype library. @Gowtham_Rao @cukarthik
Clinical Description (From OHDSI Symposium 2022 Poster #86):
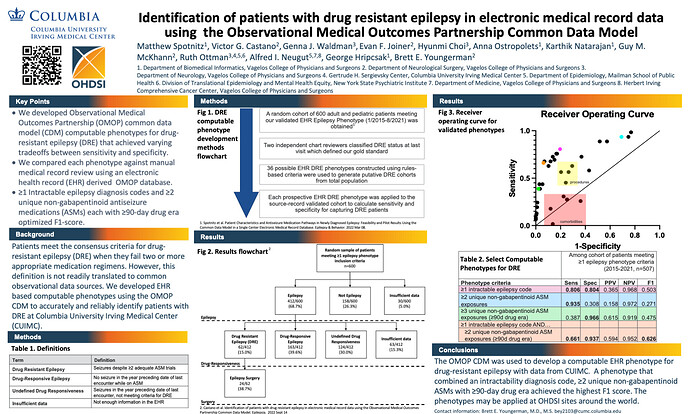
Patients meet the consensus criteria for drug-resistant epilepsy (DRE) when they fail two or more appropriate medication regimens. However, this definition is not readily translated to common observational data sources. We developed EHR based computable phenotypes using the OMOP CDM to accurately and reliably identify patients with DRE at Columbia University Irving Medical Center (CUIMC).
From the Epilepsia manuscript by Castano et. al.:
“Approximately one third of patients with epilepsy continue to have seizures despite two or more appropriate medication regimens and meet the consensus definition for drug-resistant epilepsy (DRE).” (Kwan P Epilepsia, Kwan P NEJM)
"The International League Against Epilepsy (ILAE) has defined DRE by consensus as “failure of adequate trials of two tolerated, appropriately chosen and used antiepileptic drug schedules to achieve sustained seizure freedom.” (Kwan P et. al. Epilepsia)
“With continued polypharmacy alone, the chances of achieving seizure freedom are modest, perhaps as low as 3%.”
“Patients with uncontrolled seizures are at increased risk of death, injury, cognitive decline, psychiatric illness, and decreased quality of life.”
“DRE is also associated with substantially higher relative costs of illness.”
Literature Review:
A literature review was done in preparation for the recent publication (Identification of patients with drug-resistant epilepsy in electronic medical record data using the Observational Medical Outcomes Partnership Common Data Model - PubMed). Some representative references are below
Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med . 2000; 342(5): 314– 9.
Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia . 2010; 51(6): 1069– 77.
Brodie MJ, Barry SJE, Bamagous GA, Norrie JD, Kwan P. Patterns of treatment response in newly diagnosed epilepsy. Neurology . 2012; 78(20): 1548– 54.
Sperling MR, Barshow S, Nei M, Asadi-Pooya AA. A reappraisal of mortality after epilepsy surgery. Neurology . 2016; 86(21): 1938– 44.
Jokeit H, Ebner A. Long term effects of refractory temporal lobe epilepsy on cognitive abilities: a cross sectional study. J Neurol Neurosurg Psychiatry . 1999; 67(1): 44– 50.
Spencer SS, Berg AT, Vickrey BG, Sperling MR, Bazil CW, Haut S, et al. Health-related quality of life over time since resective epilepsy surgery. Ann Neurol . 2007; 62(4): 327– 34.
Begley C, Wagner RG, Abraham A, Beghi E, Newton C, Kwon CS, et al. The global cost of epilepsy: a systematic review and extrapolation. Epilepsia . 2022; 63(4): 892– 903.
Willems LM, Hochbaum M, Frey K, Schulz J, Menzler K, Langenbruch L, et al. Multicenter, cross-sectional study of the costs of illness and cost-driving factors in adult patients with epilepsy. Epilepsia . 2022; 63(4): 904– 18.
We have tabulated the results from prior studies that have validated drug resistant epilepsy with source-level data in our publication. Here are those references:
Freedman DA, Grinspan Z, Glynn P, Mittlesteadt J, Dawes A, Patel AD. Validating coding for the identification of pediatric treatment resistant epilepsy patients. Child Neurol Open. 2021;8:2329048X211037806.
Pan S, Wu A, Weiner M, Grinspan MZ. Development and evaluation of computable phenotypes in pediatric epilepsy: 3 cases. J Child Neurol. 2021;36(11):990-7.
Hill CE, Lin CC, Terman SW, Rath S, Parent JM, Skolarus LE, et al. Definitions of drug-resistant epilepsy for administrative claims data research. Neurology. 2021;97(13):e1343-50
Logic Description:
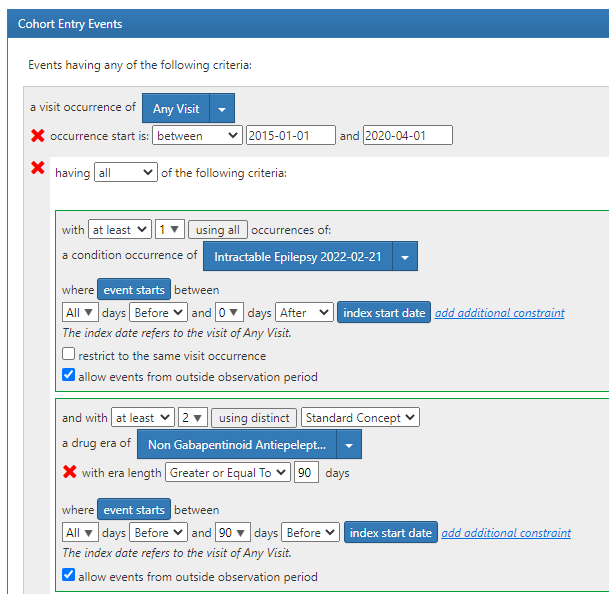
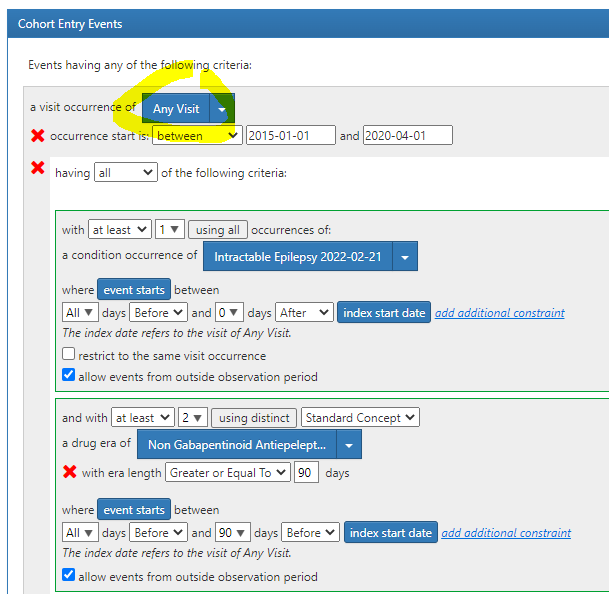
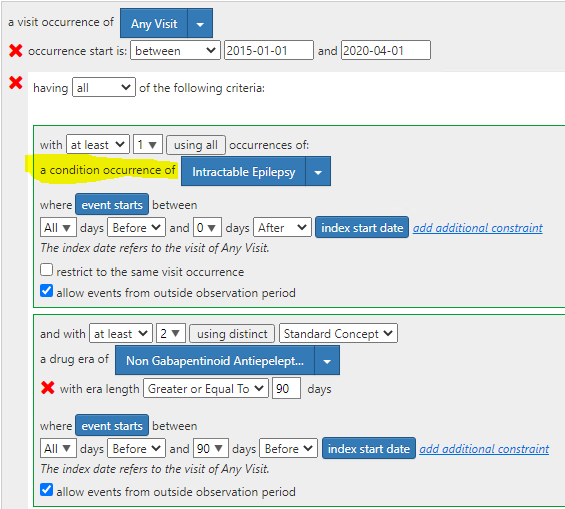
Our index event was any visit between 01/01/2015 and 04/01/2020. Additionally, patients had to have at least 1 intractable epilepsy diagnosis between all and 0 days prior to the visit and at least 2 distinct 90-day drug eras of non-gabapentinoid antiseizure medications anytime between all and 90 days prior to the index visit. We also restricted to patients who had at least 365 days of observation after the index event.
We calculated the sensitivity, specificity, PPV, NPV and F1 score of this phenotype by cross validation with a random sample of 600 epilepsy patients from a previously validated cohort. We compared the performance of this phenotype to multiple alternatives.
Among 412 patients with source record-confirmed epilepsy, 62 (15.0%) had DRE, 163 (39.6%) had drug-responsive epilepsy, 124 (30.0%) had undefined drug responsiveness, and 63 (15.3%) had insufficient records. The best performing phenotype for DRE in terms of the F1 score was the presence of ≥1 intractable epilepsy code and ≥2 unique non-gabapentinoid ASM exposures each with ≥90-day drug era (sensitivity = .661, specificity = .937, PPV = .594, NPV = .952, F1 score = .626). Several phenotypes achieved higher sensitivity at the expense of specificity and vice versa.
Overall, the F1 score was highest for this phenotype. A more extended explanation of the validation process can be found in the poster or publication.
Drug Resistant Epilepsy Phenotype.docx (67.2 KB)