Happy Saturday everyone!
It is non-small cell lung cancer (NSCLC) day.
I am presenting on behalf of a phantastic group: @dkosareva, @mgurley, @Ajit_Londhe, @CarolynB, Xerxes Pundole, and @jshaw03. And special thanks to @Patrick_Ryan.
I am going to make a little bit of a change today and start talking about what we want to define today.
Our goal today is to come up with a phenotype for NSCLC. We would like to identify patients with a diagnosis of NSCLC, irrespective of the disease stage. We are also interested at all NSCLC patients (prevalent cases) and NOT newly diagnosed (incident) cases.
I have a very long summary of the clinical definition of NSCLC but I am afraid my summary will not do a fair job in describing the disease. NSCLC is not just one disease but many each with its own characteristics, risk factors, behaviors and treatments so I could not convince myself to share the long summary here with you. This Nature Review on NSCLC gives a good overview on the disease. Here, I will focus on a broad definition of the disease as it pertains to our phenotype of interest.
What is NSCLC? NSCLC is type of cancer that originates in the lung. NCI dictionary defines NSCLC as “a group of lung cancers named for the kinds of cells found in the cancer and how the cells look under a microscope. The three main types of non-small cell lung cancer are adenocarcinoma (most common), squamous cell carcinoma, and large cell carcinoma. Non-small cell lung cancer is the most common of the two main types of lung cancer (non-small cell lung cancer and small cell lung cancer (SCLC)).”
Sounds simple and straight forward. All I need to do to find my cohort of NSCLC patients is to first identify tumors that originate in the lung (location or topology of the tumor) and then identify the subset of lung cancer patients with a specific tumor histology: namely adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. The combination of topology+histology is what is called the base diagnosis and was introduced as a part of the Cancer Diagnosis Model in the Oncology Module.
Not that easy ![]()
The ICD-9 and ICD-10 coding systems do not not distinguish between histological types. So we cannot really differentiate NSCLC from SCLC.
What does the literature tell us?
-
in 2008, Duh et al developed an algorithm to identify SCLC cases from all lung cancer in administrative claims databases. They used American Cancer Society (ACS) and National Comprehensive Cancer Network (NCCN) treatment guidelines and clinical expertise to identify SCLC. Later on, this algorithm was modified by reversing the inclusion and exclusion criteria to identify NSCLC patients in claims databases. The Modified Duh Algorithm uses chemotherapy regimens applied to patients with SCLC as exclusion criteria and first-line chemotherapy regimens administered to patients with NSCLC as inclusion criteria. Although widely used, the algorithm has not been validated.
-
In 2017, Turner et al used the existing Modified Duh Algorithm and updated to include first-line treatments and test recommendations for patients with NSCLC and SCLC according to 2015 ACS and 2016 NCCN guidelines. The validation was performed using the HealthCore Integrated Research Environment (HIRE)-Oncology clinical linked to HealthCore Integrated Research Database (HIRD). They reported a sensitivity (94.8%), specificity (81.1%), positive predictive value (PPV) (95.3%).
-
In 2020, Balzi et al proposed another algorithm to distinguish SCLC from NSCLC. They identified lung cancer patients using ICD-9CM diagnosis codes and excluded patients with other malignancy and those initiated Etoposide or Lanreotide in the first 180 days after diagnosis. They reported a sensitivity of 88.8% and a high PPV of 90.2% but a suboptimal specificity (53.7%) and NPV (50%).
In summary, the NSCLC algorithms in the literature is a combination of lung cancer diagnosis and SCLC and NSCLC treatments.
Can we use these learnings to come up with a good phenotype for NSCLC? Let’s dissect this a bit more:
-
Not all lung cancer people initiate treatment. The proportion of untreated patients with NSCLC varies by stage, ranging from 7% to 45%. This figure can be as high as 90% for older and more frail patients (David et al). Similar rates have been reported for SCLC. Using any of the proposed algorithm means that we will miss a substantial proportion of our NSCLC patients that did not receive treatment (impacting sensitivity). Remember, our intension was to build a NSCLC phenotype not a phenotype for NSCLC patients who initiated a treatment. An algorithm based on future treatment would therefore give use an unreal picture of all NSCLC patients.
-
Using treatment information to include/exclude patients can help distinguish the diseases as long as treatments are distinct. The overlap between treatments compromise specificity of the definition. Is that the case?
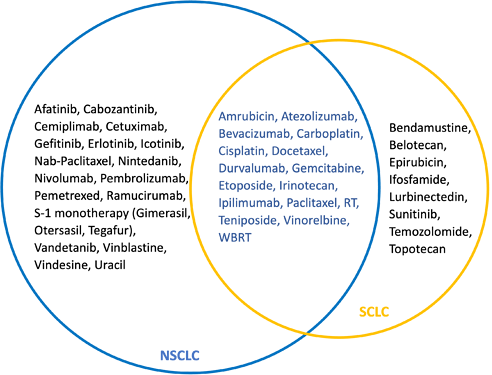
The treatment landscape of NSCLC and SCLC and clinical practice guidelines are constantly changing. Developing a universal and reproducible phenotype for NSCLC using treatment information requires regular update and maintenance to ensure the definition captures all the newly approved treatments across geographies. In addition, the performance of the definition needs to be assessed regularly. Introduction of new distinct treatments for each tumor types would lead to better specificity of the definition while approval of the same drug for both conditions will lead to a loss in specificity.How similar or different are the SCLC and NSCLC treatments? Let’s take a look at the NSCLC and SCLC treatments as described in HemOnc.
The following regimens are used for both diseases:
| Regimen | NSCLC indication(s) | SCLC indication(s) |
|---|---|---|
| Carboplatin & Etoposide (CE) | 1L for advanced or metastatic disease | LS induction, LS definitive therapy, LS adjuvant therapy, ES induction, ES relapse refractory disease |
| Carboplatin & Paclitaxel (CP) & Ipilimumab | 1L for advanced or metastatic disease | ES induction |
| Cisplatin & Etoposide (EP) | Adjuvant and neo-adjuvant therapy, 1L for advanced or metastatic disease | LS induction, LS definitve therapy, ES induction, ES relapse refractory disease |
| Cisplatin & Irinotecan (IC) | 1L for advanced or metastatic disease | LS consolidation for upfront therapy, ES induction, |
| Radiation therapy | Definitve therapy for locally advanced disease | LS consolidation for upfront therapy |
| Docetaxel monotherapy | 1L for advanced or metastatic disease (elderly or poor performance status), maintenance after 1st, 2nd line, 3rd line and subsequent lines of therapy | ES induction |
| Gemcitabine monotherapy | 1L for advanced or metastatic disease (elderly or poor performance status), maintenance after 1st, 3rd line and subsequent lines of therapy | Relapse refractory disease |
| Bevacizumab monotherapy | Maintenance therapy after 1st line | ES maintenance |
| Ipilimumab monotherapy | Maintenance therapy after 1st line | ES maintenance |
| Amrubicin monotherapy | Advanced or metastatic disease, subsequent lines of therapy | Relapse refractory disease |
| Cisplatin, Etoposide, RT | Definitive therapy for locally advanced disease | LS definitive therapy |
So technically, we cannot differentiate NSCLC and SCLC for patients on any of these regimens. We can infer the disease based on the treatment and our prior knowledge from their frequency of their use in either patient population (mmm sounds like a lot of guesswork and not really reproducible ![]() ). Also, the impact of treatment overlap on the specificity and accuracy of the definition depends on the how frequent these treatments are being used in each setting and as mentioned above, this can easily change over time. But similar to Turner et al, we can create a phenotype based on a combination of LC, SCLC treatment and NSCLC treatment: lung cancer patients WITH NSCLC treatment AND NOT SCLC treatment. We can test it out and see what the data tells us.
). Also, the impact of treatment overlap on the specificity and accuracy of the definition depends on the how frequent these treatments are being used in each setting and as mentioned above, this can easily change over time. But similar to Turner et al, we can create a phenotype based on a combination of LC, SCLC treatment and NSCLC treatment: lung cancer patients WITH NSCLC treatment AND NOT SCLC treatment. We can test it out and see what the data tells us.
But wait! there is another problem here: Most cancer treatments are administered in chemotherapy regimens with complex dosing and scheduling in multiple cycles and are often combined with targeted therapies, immunotherapies, surgery or radiotherapy. We do not have that in the data to use in for our definition. what we have is a list of individual drugs or procedures, basically the components of the regimens. There are several ongoing efforts in the community to abstract treatment regimens from different combinations of drugs and procedures. But until then, we are limited to the list of drugs and procedures. Ok! let’s take another look and do a quick comparison between NSCLC and SCLC treatments:
There seems to be a substantial overlap between drugs used for treatment of the two conditions and some of which are commonly used in both conditions (as a part of different regimens).
-
Using treatment to accurately distinguish SCLC and NSCLC treated populations depends on the completeness of treatment information in our datasources. For example, oncology EMR data coming from oncology clinics (a common source of data used for observational oncology research) have a good capture of systemic antineoplastic use but lack information on surgery, radiotherapy, and other inpatient procedures. Similarly Claims are limited in capturing inpatient administration. Each datasource is showing us one snapshot and a different view of the patients treatment journey and not all. Can we rely on these limited snapshots to get to our NSCLC patients?
-
What about multiple primaries? Up to 20% of lung cancer patients develop a secondary tumor and the most common secondary malignancy in patients with lung cancer is in the lung: ~57% of patients with SCLCL develop NSCLC. Additionally, a history of previous malignancy in patients with lung cancer is reported in ~11% of cases. Having multiple primary tumors complicate the use of treatment for identification of NSCLC patients. But if we exclude them, our definition will only identify a subset of all NSCLC patients with no other primaries who initiated treatment.
-
A definition based on treatment uses future events (here, treatment) to define the patients population. This means we will only capture a subset of patients who survived and received a treatment. Sounds like immortal time (AVOID IT BY ALL MEANS). If I want to describe use this definition to do a characterization of NSCLC patients and their outcomes after diagnosis of NSCLC, I will be looking at the subset who have survived and are being treated (selection bias). If I want to describe treatments patterns, I am basically describing what I have used to define my cohort. I cannot see any off-label use or disparities in treatment. The definition would work if I was interested in NSCLC treated patients as a phenotype and correctly identify the index date but that was not what we intended to create.
How should we move forward?
-
Do not create a phenotype in the presence of this much uncertainty. Instead, let’s work together to enrich the data either through linkage to tumor registry or other sources with information on tumor histology and other necessary attributes or by looking into the notes. While we strongly recommend and encourage the community to join the oncology WG and others in enhancing oncology data, we still want to comprehensively assess the limitations and drawback of the current data in defining NSCLC.
-
Create the following phenotypes:
- Probably NSCLC treated cohort: Lung cancer diagnosis AND NSCLC specific treatment initiation
- Probably SCLC treated cohort:Lung cancer diagnosis AND SCLC specific treatment initiation
- We have no clue lung cancer cohort: Lung cancer diagnosis AND initiation of treatments commonly used in both conditions
We will look at the cohort diagnostics results and also do a simple characterization to further understand the attrition based on different treatments and potential differences in the patient population.
-
Create a definition for NSCLC and SCLC following the Oncology Module recommendation from a combination of histology and topology. We will use these definitions (in addition to the previous ones) on a couple of our oncology data sources to investigate the potential overlap and the degree of misclassification associated with using treatments for defining the cohorts.