First I want to thank everyone for the informative phenotype discussions so far. I have learned a lot from reading them.
Along with lots of help from my colleagues Darya Kosareva and @agolozar I have created a few cohort definitions for triple negative breast cancer to share. In the process I have learned some lessons about the importance of understanding the vocabulary and how clinical concepts are recorded in actual data. I’d like to summarize the cohorts first then put the clinical definition and references in then next post. Keep in mind that I rely on others for clinical expertise since I have no clinical training and have been lucky to have many people around me with significant clinical expertise.
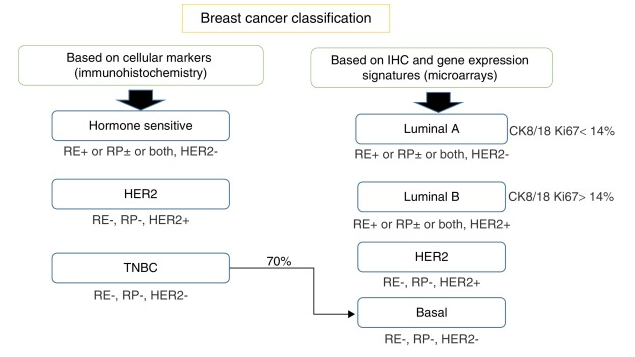
Triple negative breast cancer (TNBC) is an aggressive form of breast cancer that is estrogen receptor (ER), progesterone receptor (PR) negative, and HER2 negative. There are few treatment options and the prognosis is bad. I’m trying to identify incident cases of TNBC. I’ll put a more complete clinical description in the next post.
Initially I expected standardization to allow for the creation of one cohort definition that captures persons meeting a particular phenotype definition across a network of databases without having direct access to the data or knowing how the source data was recorded. This expectation is based on two assumptions.
- We have a standard vocabulary where each clinical idea has a unique representation
- Source data is accurately mapped into that standard representation
There is a lot to unpack in those two assumptions. (e.g. what do I mean by ‘accurately mapped’). Putting that aside for a moment I started with a naively simple cohort definition that I think represents how some OMOP users would like to use a standardized clinical database. TNBC is very clearly a condition. When searching the vocabulary do indeed find a single standard concept for Triple-negative breast cancer (45768522). I might expect that this concept would provide good sensitivity and specificity for the TNBC patients in any OMOP database. So the simplest possible cohort definition that I’d expect to be entirely database agnostic is based on this single condition concept.
Triple negative breast cancer (single diagnosis)
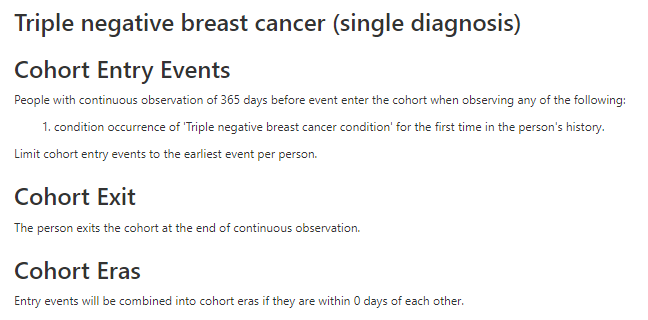
This definition is problematic for a few reasons. One issue with 45768522 is that no ICD10 codes are mapped to it (because I don’t think there are any specific triple negative ICD10 codes) so this code isn’t really used in OHDSI databases and will miss many/most TNBC patients. PHEOBE shows only about 1700 records across OHDSI with concept ID 45768522 . We need to use a more general clinical concept of “primary malignant breast cancer” and then narrow in on TNBC using measurements (ER/PR/HER2). This requires our data to have measurement data which excludes some claims databases. We also have to decide whether to exclude breast cancer patients who are clearly not triple negative or include patients who have negative test results for ER/PR/HER2. When we started looking into these tests we found many relevant measurement concepts. Some of them are specific to one of ER/PR/HER2 while others are a panel that include all three. We also have the option of using measurement values. OMOP has value concepts for ER negative, ER positive, PR negative, PR positive, HER 2 negative, HER2 positive as well as a measurement value for triple negative. However as Christian points out below these are legacy concepts and will be deprecated soon so we should also include the new OMOP Genomic concepts. However if we require the new measurement concepts to be present we will lose patients in databases that are not using these measurement concepts yet. In short it gets complicated quickly.
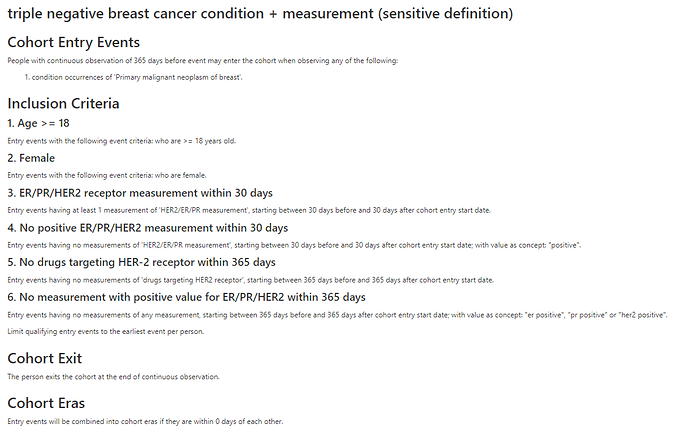
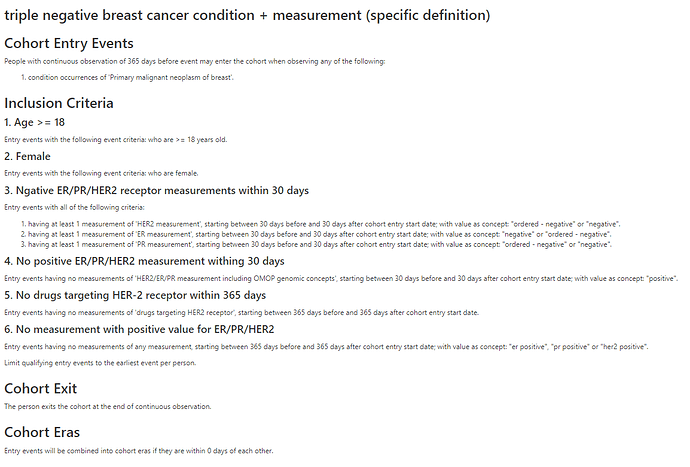
We created a sensitive (exclusion based) cohort and an specific (inclusion based) cohort using a general condition concept and measurements/measurement values. We also used HER2 targeting drugs to further exclude non triple negative patients.
triple negative breast cancer condition + measurement (sensitive definition)
triple negative breast cancer condition + measurement (specific definition)
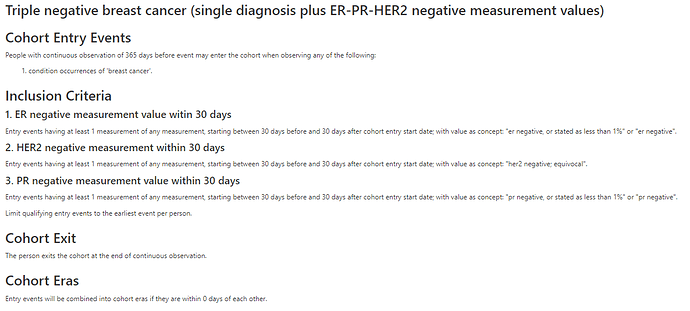
We also have measurement values we can use so I tried creating another specific cohort based on the occurrence of ER-, PR-, and HER2- measurement values alone. I tried another cohort using just the the combined triple negative measurement value.
Triple negative breast cancer (single diagnosis plus ER-PR-HER2 negative measurement values)
Triple negative breast cancer (single diagnosis plus single ER-PR-HER2 negative measurement value)
I think that these might work on some databases but I also believe I have only scratched the surface of this phenotype definition. I didn’t get into the is the issue of finding the correct index data described in this poster but I think requiring an ER/PR/HER2 measurement within some time frame around the diagnosis helps to anchor on the correct incident index date. Some of the published algorithms we looked required two diagnoses at least X days but no more than Y days apart to exclude rule out diagnoses. This logic makes sense for claims and EHR databases but not for registry databases.
The main takeaway for me is that phenotyping is not database agnostic. There are many possible approaches and the best one depends depends on the type of source data and how it was mapped. I’m not sure it is possible to develop a single rule based TNBC phenotype algorithm in Atlas that works well on any OMOP database. Phenotyping algorithms sit between ETL and standardized analytics where ETL is entirely database specific and standardized analyses are database agnostic. I’m glad the community is building a library of phenotypes for us to use in standardized analytics!


 .
.