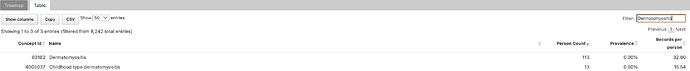
Good afternoon, we are happy to announce an upcoming network study lead by the Johns Hopkins Myositis Center in Baltimore, MD, USA. The goal of the study is to evaluate several phenotypes for a rare rheumatic disease known as ‘dermatomyositis’. We intend to characterize the performance of each phenotype using PheValuator on a variety of data sources along with comparison to a gold standard chart review of our own patient population.
Corresponding study lead: Christopher Mecoli, MD, MPH; cmecoli1@jhmi.edu @Christopher_Mecoli @benwmar @wkelly19
GitHub: https://github.com/ohdsi-studies/DermatomyositisPhenotypeCharacterization
Protocol: In development
Infrastructure and Data: The ability to execute PheValuator and DatabaseDiagnostics on a CDM likely to contain adult patients with dermatomyositis. The CDM can include inpatient, outpatient, EHR, claims, registry, or any combination thereof.
Participant deadline: If you are interested in participating in the study, please contact the study team by May 15th.
Primary and secondary objectives:
This study’s main objective is to evaluate and validate different dermatomyositis phenotypes across a range of OMOP databases. Secondary objectives include (a) Validating PheValuator’s probabilistic gold standard against a manual chart review gold standard in an EMR-based OMOP CDM; (b) To characterize index misclassification errors in the EMR based CDM of a tertiary referral center; (c) Raise awareness of OHDSI and the OMOP CDM in the clinical rheumatology community; and (d) Provide proof of concept of potential to perform large-scale network studies in rare diseases.
Background and rationale:
Rare diseases like dermatomyositis (DM) present a greater challenge in observational research studies, where the execution of large-scale studies is even more important in generating evidence due to the sparse cases observed in real-word data. This study presents the development and evaluation of a computable phenotype for DM to support coordinated analysis of this rare disease across manifold data sources while addressing reproducibility and generalizability challenges in the generation of real-world evidence supporting care for DM.
Additionally, within the rheumatology clinical community, publications addressing OHDSI methods have historically been limited to informatics journals not typically followed by clinical investigators. By presenting the PheValuator probabilistic gold standard alongside classification via manual chart review conducted at the Johns Hopkins Myositis Center, we aim to publish in a clinical journal and introduce OHDSI methodologies to a wider clinical audience in the field of rheumatology.