It’s not a function in atlas, but we’ve written custom sql to generate person-level episodes based on age criteria:
\
with ages as (
SELECT 2 as age_id, 0 as age_low, 5 as age_high
UNION
SELECT 3 as age_id, 6 as age_low, 17 as age_high
UNION
SELECT 4 as age_id, 18 as age_low, 34 as age_high
UNION
SELECT 5 as age_id, 35 as age_low, 54 as age_high
UNION
SELECT 6 as age_id, 55 as age_low, 64 as age_high
UNION
SELECT 7 as age_id, 65 as age_low, 74 as age_high
UNION
SELECT 8 as age_id, 75 as age_low, 84 as age_high
UNION
SELECT 9 as age_id, 85 as age_low, 114 as age_high
),
genders as (
SELECT 1 as gender_id, 8532 as gender_concept_id, 'Female' as gender_name
UNION
SELECT 2 as gender_id, 8507 as gender_concept_id, 'Male' as gender_name
)
SELECT ages.age_id*10+genders.gender_id as subgroup_id, age_low, age_high, gender_concept_id, gender_name
INTO #subgroups
FROM ages, genders
;
INSERT INTO @target_cohort_table (cohort_definition_id, subject_id, cohort_start_date, cohort_end_date)
SELECT s1.subgroup_id AS cohort_definition_id, op1.person_id AS subject_id,
CASE WHEN YEAR(op1.observation_period_start_date) - p1.year_of_birth >= s1.age_low
THEN op1.observation_period_start_date
ELSE DATEFROMPARTS(p1.year_of_birth + s1.age_low,1,1) END
AS cohort_start_date,
CASE WHEN YEAR(op1.observation_period_end_date) - p1.year_of_birth <= s1.age_high
THEN op1.observation_period_end_date
ELSE DATEFROMPARTS(p1.year_of_birth + s1.age_high,12,31) END
AS cohort_end_date
FROM @cdm_database_schema.observation_period op1
INNER JOIN @cdm_database_schema.person p1 ON op1.person_id = p1.person_id
INNER JOIN #subgroups s1 ON DATEFROMPARTS(p1.year_of_birth + s1.age_low,1,1) <= op1.observation_period_end_date
AND DATEFROMPARTS(p1.year_of_birth + s1.age_high,12,31) >= op1.observation_period_start_date
AND p1.gender_concept_id = s1.gender_concept_id
;
drop table #subgroups;
This is in OHDSI-Sql syntax, so you’d have to render and translate this sql using SqlRender.
You can run this query on your own CDM, and you’ll get a set of cohort records (if you just run the SELECT at the end) that gives you each person’s membership in each age/gender category that’s defined in the temp table #subgroups. I would use a similar query as this to define a ‘demograpic era’ criteria for building entry events based on age and gender for a period of time.
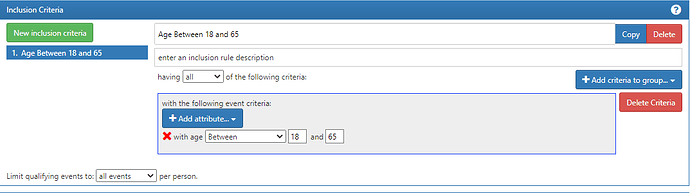
That is correct except there’s a ‘bug/feature’ in circe that may catch you unaware: when using ‘age’ criteria, it checks the age based on the record’s start date (ie: the observation_period_start_date) and not the user-defined start date.
The fix is easy, tho: move that logic to an inclusion rule like this:
What this does is it takes each entry event (which you made as Jan 1 of each year) and a person will only be included in he cohort at that date if they are in the right age. The reason why it works is that the inclusion rule will work off the dates yielded by the entry events (which you defined the start and end dates) vs. if you use the criteria directly on the observation period criteria, age criteria is based off the start_date of the record.