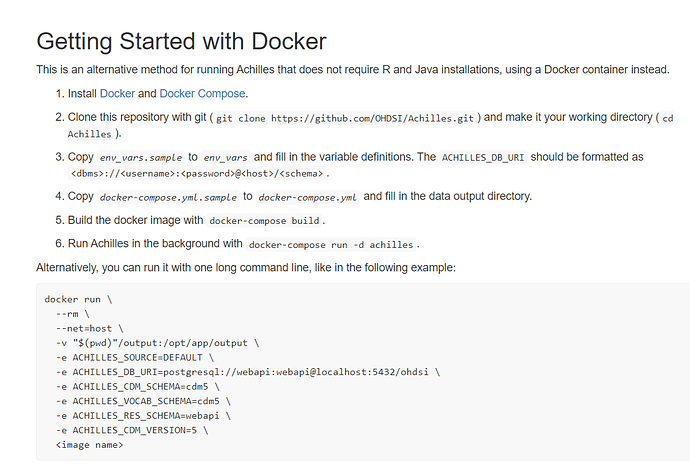
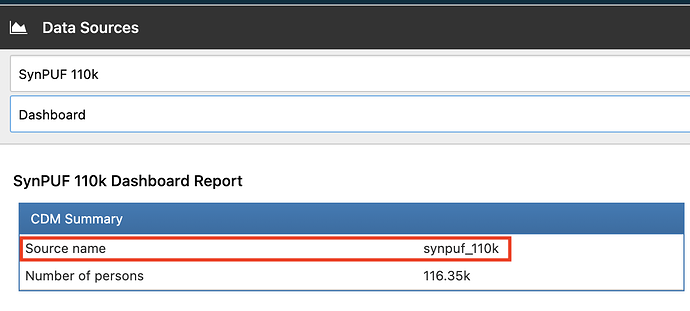
Let me see if I understand some instructions. Since I am using postgres I need to use:
library(Achilles)
connectionDetails <- createConnectionDetails(
dbms="postgres",
server="mydata.server.edu",
user="secret",
password='secret',
port="5432")
In my case, the Atlas server is: https://hsc-ctsc-atlas.health.unm.edu/
where:
Source Daimons
Results atlas_results
Temp atlas_temp
Vocabulary mdcr2003_2016
CDM mdcr2003_2016
CEM
CEMResults
In my Atlas the source name is Truven.
SingleThreading:
achilles(connectionDetails,
cdmDatabaseSchema = "mdcr2003_2016",
resultsDatabaseSchema="atlas_results",
vocabDatabaseSchema = "mdcr2003_2016",
numThreads = 1,
sourceName = "Truven",
cdmVersion = "5.3.0",
runHeel = TRUE,
runCostAnalysis = TRUE)
Fur Multithreading:
achilles(connectionDetails,
cdmDatabaseSchema = " mdcr2003_2016",
resultsDatabaseSchema = "atlas_results",
scratchDatabaseSchema = "scratch",
vocabDatabaseSchema = "mdcr2003_2016",
numThreads = 10,
sourceName = "Truven",
cdmVersion = "5.3.0",
runHeel = TRUE,
runCostAnalysis = TRUE)
Is this syntax correct? as far as I know we do not have the scratch schema.
Thanks again