Team:
Five years ago, a few of us thought it would be fun to come together and create an open science community and wondered aloud if anyone would join us. Each of us had been working in our own separate silos, some of us had collaborated on OMOP, but all of us shared a basic belief that real-world evidence could dramatically improve healthcare if done right and none of us believed we could solve this problem by ourselves. Our idea was pretty simple: create an open place where all people from all backgrounds and perspectives felt encouraged to collaborate on the research, development, and application of analytic solutions for observational data to generate and disseminate evidence that could inform medical decision-making. So, we started OHDSI, curious where the journey may lead us.
Fast forward 5 years, and I could have never anticipated how fast and large we’ve seen our community grow. On this forum alone, we now have 2,455 users engaging in discussions about how to use their observational data to generate reliable real-world evidence. For last year’s OHDSI US Symposium, we asked the community to self-identify what observational data sources have you converted to the OMOP common data model, and we found 97 databases in 19 countries, collectively representing more than 2 billion patient records. At last month’s OHDSI Europe symposium, we had 260 people from 27 different countries come together to collaborate. Just last week, we announced the opening of registration for the OHDSI US Symposium, which will take place in DC in September, and already 135 of the 500 available seats have been taken by researchers from government, academia, industry, and health systems. OHDSI events have been hosted in New York and Atlanta and San Francisco and Rotterdam and Oxford and Suwon and Guanzhou and Shanghai. The collective expertise and energy to work together toward a common mission: ‘to improve health by empowering a community to collaboratively generate the evidence that promotes better health decisions and better care’ to truly inspiring.
Today, we have a vibrant community that is establishing open community standards for harmonizing the structure, content, and semantics across disparate observational databases around the world. Our community is leading cutting-edge methodological research that is identifying and evaluating scientific best practices for clinical characterization, population-level effect estimation, and patient-level prediction. We’ve applied these best practices to important clinical questions that matter to patients, whether it be specific targeted questions like the safety of levetiracetam, or the comparative effects on HbA1c amongst diabetes treatments, or large-scale analyses like the LEGEND studies for depression and hypertension. And we’re supporting national and international efforts to scale the public health impact that observational data can make, through programs such as FDA/BEST, NIH/AllOfUs, and IMI/EHDEN.
I think a lot of the tremendous progress we’ve made can be together can be attributed to our commitment to working as an open science community, and sharing in the development of open-source tools that make it possible for all of us to adopt open community data standards and apply open standardized analytics to our data. We’ve seen this commitment manifest in a robust ecosystem of open-source solutions that support the entire analytics lifecycle from data management and quality assessment to back-end packages for large-scale statistical modeling to front-end web applications for study design and execution. This ecosystem makes it possible for a researcher to generate reliable real-world evidence from their observational data with higher quality and greater transparency, and to do so in a matter of minutes what previously took months. The OHDSI ecosystem has grown to now include 148 Github repositories, with contributions by more than 100 developers.
But with any growth can come some growing pains, and supposedly when we get older, we’re supposed to become a little wiser and more mature as well. What we could get away with as a precocious upstart might be different from what we expect of ourselves as our community evolves and the user adoption of the OHDSI ecosystem expands.
Now I am not a software developer. I enjoy hacking a little code, and really like solving hard problems by rapidly prototyping ‘good enough’ solutions. I know many in the community falls into my same phenotype in that regard. But I recognize that, while the uncommented spaghetti code I write for myself may be good enough for my bespoke purpose in the heat of the moment, it is not nearly good enough for a sustainable solution that others will rely on to meet their needs. In fact, in my zest to advance science by introducing my half-baked approaches to the fold, I may be doing more harm than good. Contributing as a community developer can be challenging, because it requires being sensitive to the community ecosystem and its users. In order to build trust, a proper open science community solution is an open-source tool that is not only fully transparent and publicly available, but also one which is developed by a competent team of individuals, is fully compliant with the community standards, produces consistent results, is appropriately documented and properly tested, and is responsive to the needs of the community.
To date, we’ve been fairly inconsistent in how the community has managed its open-source development across the OHDSI ecosystem. We’ve got away with it in some areas, in part because we had a smaller team of developers who aligned on a shared understanding of best practices and faithfully followed these implicit expectations. For example, @schuemie and @msuchard have a shared vision about best practices for R package development that they have applied throughout the OHDSI methods library, and @jennareps and @Rijnbeek largely adopted those principles in the development of PatientLevelPrediction. Some aspects of these developer best practices have been documented, such as recommendations for unit tests in R or code styleguides for SQL, but other aspects are either insufficiently documented or inadequately enforced. When ATLAS was in its infancy, @Frank, @Chris_Knoll and @anthonysena could grok the whole codebase on their own with little help from others. But now that ATLAS has evolved to become a unifying platform to support the design and execution of observational studies, with a myriad of analytic features and is comprised of community contributions from dozens of developers, its difficult to imagine that anyone could get their hands around all the details of everyone component. And concurrent with the growth of complexity of the tools has come the growth of the user community; because the OHDSI ecosystem is now actively deployed at so many institutions, it is increasingly important for us to have software releases of each of its components that the community can trust.
To meet these emerging community needs, I propose that we formalize our open community development processes to align on shared expectations and to hold ourselves accountable to the standards we expect for our community. Before we launched OHDSI, I read Producing Open Source Software by Karl Fogel, and in thinking through the issues our community currently faces, I found myself going back to this resource and finding it still quite useful to think through what makes most sense for our community. One insight that is particularly apparent is that there is no one ‘right way’ to build a open source solution, but it is very important that a community aligns to the ‘right way for them’, whatever that may be. So with that, I offer the following strawman to stimulate the community discussion:
I recommend that, for each open-source product with the OHDSI ecosystem, that we have a named product owner. The project owner assumes the following responsibilities:
- Serve as a benevolent dictator, with final decision-making authority on the project
- Ensure project codebase remains open source and publicly available through OHDSI GitHub repository (Apache 2.0 license for open community use)
- Produce documentation that details desired objective and available features, installation instructions (including system requirement)
- Ensure an adequate test framework is properly implemented prior to any release. Testing should be fully described as part of the release.
- Establish and communicate a project release process and roadmap for future development, which is responsive to user community and contributors
- Coordinate with other project owners who have logical dependencies on the project
- Support transition of the project to a new project owner when necessary
I recommend that we also align on shared expectations for our contributor community:
- Use the tools
- Support documentation and facilitate adoption by community
- Identify bugs and post them on the GitHub issue tracker
- Propose solutions to any identified bugs to the project owner for their consideration
- Suggest potential feature enhancements on the GitHub issue tracker or OHDSI forums
- Collaborate with project owner to design and implement new capabilities
I recommend that the OHDSI Coordinating Center, led by @hripcsa, continue to oversee the OHDSI community assets, including the OHDSI Github repository, and work with each project owner to ensure that each project satisfies the recommended community standards. If a project does not conform to agreed standards, the OHDSI coordinating center reserves the right to tag the project as non-compliant. If there is disagreement in the community about ongoing project development activities, the OHDSI coordinating center will serve as the final arbitrator.
If someone wants to create a new project to add to the OHDSI ecosystem, they can contact me and I will be happy to discuss and coordinate with the other project owners to initiate the effort (including creating the OHDSI Github repository).
Below is my recommendation for named project owners for components within the current OHDSI ecosystem:
| OHDSI Ecosystem Project owners | |
|---|---|
| Common Data Model | |
| CommonDataModel | Clair Blacketer |
| DDLgenerator | Clair Blacketer |
| Standardized Vocabularies | |
| Vocabulary-v5.0 | Christian Reich |
| ATHENA | Greg Klebanov |
| Tantulus | Peter Rijnbeek |
| ATLAS platform | |
| User experience | Patrick Ryan |
| Technical integration | Greg Klebanov |
| Security | Pavel Grafkin |
| Data sources | Ajit Londhe |
| Vocabulary search | Frank DeFalco |
| Cohort definition | Chris Knoll |
| Characterization | Pavel Grafkin |
| Pathways | Chris Knoll |
| Incidence | Chris Knoll |
| Estimation | Anthony Sena |
| Prediction | Anthony Sena |
| ARACHNE | Greg Klebanov |
| ACHILLES | Ajit Londhe |
| OHDSI Methods library | |
| Population-level estimation | |
| CohortMethod | Martijn Schuemie |
| SelfControlledCaseSeries | Martijn Schuemie |
| SelfControlledCohort | Martijn Schuemie |
| CaseControl | Martijn Schuemie |
| CaseCrossover | Martijn Schuemie |
| PatientLevelPrediction | Jenna Reps |
| BigKnn | Martijn Schuemie |
| Methods characterization | |
| EmpiricalCalibration | Martijn Schuemie |
| MethodEvaluation | Martijn Schuemie |
| EvidenceSynthesis | Martijn Schuemie |
| Supporting Methods Packages | |
| Cyclops | Marc Suchard |
| DatabaseConnector | Martijn Schuemie |
| SqlRender | Martijn Schuemie |
| ParallelLogger | Martijn Schuemie |
| FeatureExtraction | Martijn Schuemie |
| OhdsiRTools | Martijn Schuemie |
| Hydra | Martijn Schuemie |
| drat | Marc Suchard |
| LEGEND | Martijn Schuemie |
| ETL support tools | |
| WhiteRabbit | Martijn Schuemie |
| Usagi | Martijn Schuemie |
| Other community solutions: | |
| PhenotypeLibrary | Aaron Potvien |
| PheValuator | Joel Swerdel |
| Aphrodite | Juan Banda |
| Criteria2Query | Chi Yuan |
| QueryLibrary | Peter Rijnbeek |
| OHDSIonAWS | James Wiggins |
| Broadsea | Lee Evans |
| BookOfOhdsi | David Madigan |
| Aegis | Chan You |
| ShinyDeploy | Lee Evans |
| Circe | Chris Knoll |
| CommonEvidenceModel | Erica Voss |
| THEMIS | Mui Van Zandt |
Below is a rough schematic of the interplay between these various projects within the OHDSI ecosystem, as best as I can see it. I expect this will be an evolving landscape, but already it should illustrate while maturing our processes is likely a good idea, given the complexity and interdependencies between our various components.
I welcome a healthy discussion from our community about how we should evolve our open community development activities. I know @pbiondich probably has the most experience of anyone in our community through his work with OpenMRS and hope that he can shed some light on this proposed direction, good, bad or indifferent. My opinions are largely informed by ongoing discussions with @Christian_Reich, @schuemie, @msuchard, and @jon_duke, but I don’t presume to speak for them. I suspect @JamesSWiggins can share some valuable insight based on the work he’s leading at AWS. I expect @gregk, @pavgra, and @lee_evans will have plenty to say as well. I look forward to hearing from you all, learning from your experience and perspectives, and working together toward developing innovative solutions that can improve health through the evidence we can collaboratively generate. We’re all on this journey together, and I’m excited for the path ahead.
Cheers,
Patrick
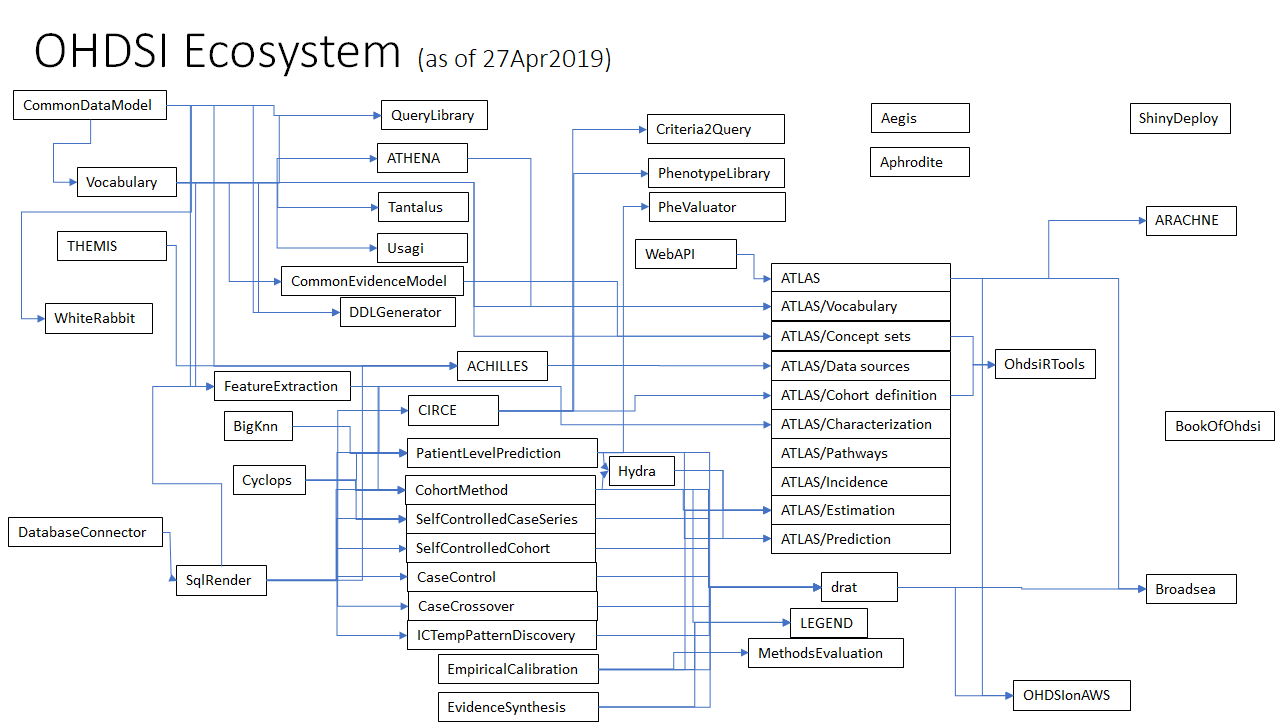
 The ways that other communities have dealt with this is via both process tweaks and governance tweaks.
The ways that other communities have dealt with this is via both process tweaks and governance tweaks.
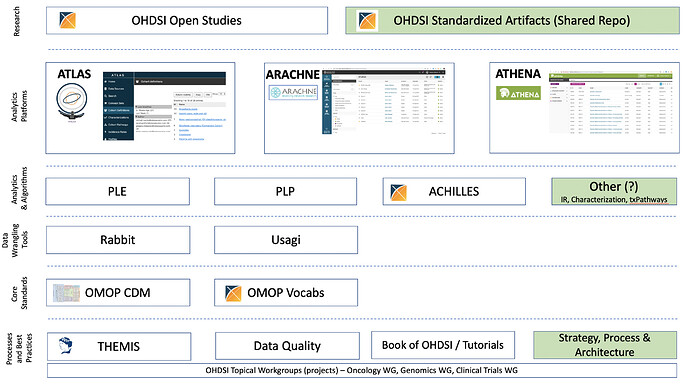
 Especially the Rabbits and Usagi. But one thing to keep in mind is that the role of ‘owner’ is a long-term commitment, so we need to make sure we have the (financial) incentives in place to allow folks to make such commitments.
Especially the Rabbits and Usagi. But one thing to keep in mind is that the role of ‘owner’ is a long-term commitment, so we need to make sure we have the (financial) incentives in place to allow folks to make such commitments.