Hello OHDSI community,
Today, I want to discuss a known but perhaps less spoken about problem, inspired by the recent forum post from @Chris_Knoll (Using the 'Mapped' function of concept set expressions).
I am a big fan of the OHDSI Cohort Definition model (i.e., the CIRCE model) and the ATLAS UI for cohort definition as it provides an elegant framework for defining entry events, inclusion rules, and exit criteria. However, I often find myself not appreciating the limitations of its concept set expression, the structure of the OMOP vocabulary tables (concept_ancestor and concept_relationship), and the sometimes-painfully ambigious distinctions between standard vs. non-standard concepts.
I believe we must improve these foundational elements. We need to reduce the inherent ‘difficulty’ in phenotyping, minimize the need for ‘vocabulary SQL acrobatics’, and escape the recurring ‘version nightmares’.
1. Complex Concept Sets are Unavoidable
Let’s start with an obvious point: we cannot avoid using complex concept set expressions. Relying solely on a conceptId plus its descendants is rarely sufficient (Sorry, @Christian_Reich ). This is because of the polyhierarchy in concept_ancestor, simply pulling in all descendants often introduces specificity errors. Lets call this established and not debate this.
2. CIRCE Concept Set Expression does not support the concept_relationship thus ignores laterality
If we agree that complex expressions are necessary, the next question is: Do our tools support building them effectively, or are we forced into complex acrobatics?
Currently, the concept set expression model in CIRCE supports only two modes of traversal:
- Hierarchy (Vertical): Using
includeDescendants, which queries the pre-computedconcept_ancestortable. This is fast and powerful for parent-child relationships. - Mapping (Limited Horizontal): Using
includeMapped. This is an occasionally used acrobatic maneuver strictly limited to the hardcoded'Maps to'relationship inconcept_relationship(as discussed in the forum link above from @Chris_Knoll ).
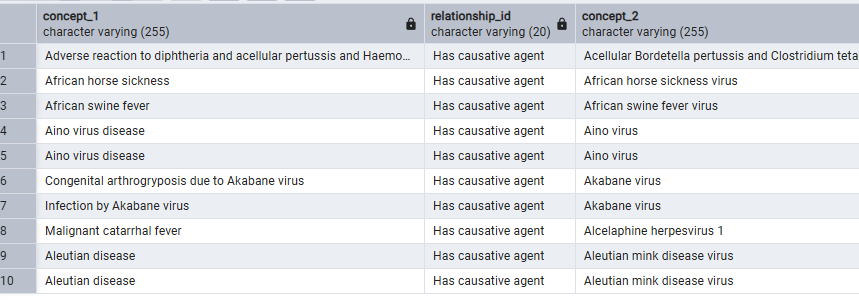
This limitation means that the rich, ‘lateral’ ontological relationships stored in concept_relationship—such as ‘Has causative agent’, ‘Has finding site’, or ‘Has active ingredient’—are essentially locked away from us within the ATLAS/CIRCE framework. We cannot use them to define our concept sets.
3. Example: The Challenge of DILI (Drug-Induced Liver Injury)
Complex phenotypes like DILI perfectly illustrate this difficulty. The clinical idea is straightforward:
Find conditions representing liver injury that are NOT caused by other specific etiologies like viruses, alcohol, or physical obstruction.
Currently, we solve this using cohort definition acrobatics—perhaps 10 inclusion rules requiring exactly 0 occurrences of hepatic viruses within -7 to +7 days of the hepatitis event, etc. We do this because the entry event concept set cannot leverage laterality; it can only navigate parent-child relationships.
What we should be able to ask the vocabulary is: “Show me all descendants of ‘Liver Injury’ that DO NOT have a ‘Caused by’ relationship to a descendant of ‘Virus’ or ‘Alcohol’.”
Because CIRCE concept set expression cannot traverse the ‘Caused by’ relationship, we are forced into a painful, manual workaround:
- Manual Curation: We must painstakingly search the vocabulary for every pre-coordinated concept that implies a non-drug cause (e.g., “Alcoholic hepatitis,” “Acute viral hepatitis B,” “Obstructive biliary disease”).
- Brittle Exclusion List: We then add this long, manually curated list to the “Excluded Concepts” panel. Its brittle because the concept set expression does not automatically add any updates to our clinical idea on what may be causing the disease.
This workaround is not just tedious; it’s the primary source of the “difficulty” and “version nightmares” I mentioned. It is error-prone (leading to low recall or specificity errors), and the list becomes obsolete with the next vocabulary update, requiring the entire manual process to be repeated. And do not get me started on the added complexity of non-standard ICD10CM/ICD9CM code mapping.
4. Why This Matters for the Future - because the community is solving this phenotyping is difficult problem.
I am hearing about many innovations being built on top of the existing concept_ancestor structure and the current CIRCE model—for example, using generative AI or vector embedding-based semantic search to build concept set expressions.
Perhaps these approaches will succeed, and we can continue to rely solely on concept_ancestor without using the concept_relationship. But perhaps we are overlooking a solution that already exists within our ontology (i.e., laterality) and hamstringing these new tools by building them on an incomplete foundation.
This is even more critical in domains like cancer phenotyping, where laterality is essential for distinguishing key characteristics:
- Origin/Status: Primary (originating in the lung) vs. Secondary (metastasis from elsewhere).
- Morphology: Malignant (cancerous) vs. Benign.
- Histology: Non-Small Cell Lung Cancer (NSCLC) vs. Small Cell Lung Cancer (SCLC).
- Location: Lower respiratory tract (lung/bronchus) vs. Upper respiratory tract.
To achieve robust, reproducible, and scalable phenotyping, we need tools that can navigate the full richness of our vocabulary, not just the hierarchy.
There are many ways to tackle this problem - if we agree this is a problem. Maybe we should dream big - rethink this completely and adopt labeled property knowledge graphs and throw away the concept_ancestor and concept_relationship (making us AI/LLM ready). Maybe we should not blindly put any concept that does not have equivalence as non-standard (e.g. ICD10CM codes that do not have 1:1 equivalence mapping) and do lossy mapping but instead make them snomed extensions and make them standard with proper descendants. Maybe we should do a breaking change to circe to support knoweldge graphs instead of SQL.