Hi Paul,
Thank you for your follow-up questions. I’ve shared a few of my thoughts below, though for some points I think it would be valuable to also hear perspectives from others in the imaging WG.
1. Linking condition_occurrence to image_feature
It is technically possible to link a condition_occurrence record to an image_feature in the same way as with measurement or observation, if the condition can be clearly tied to a specific image series (in DICOM’s hierarchy: Person → Study → Series → Instance, where the MI-CDM image_occurrence table’s basic unit is the Series).
For findings derived solely from image interpretation (including negative findings), it will be better stored in measurement or observation, while confirmed diagnoses in condition_occurrence could be linked to image_occurrence at a higher level (e.g., via the visit or episode table).
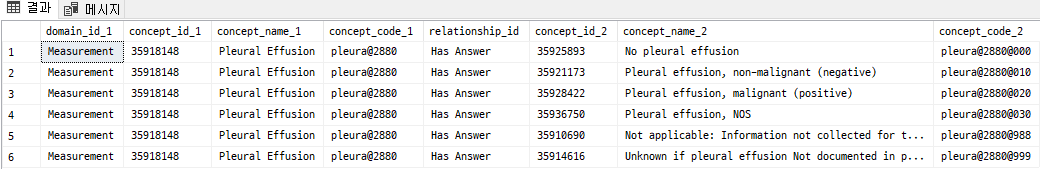
2. Vocabulary Selection
The NAACCR and LOINC examples are oncology- or test-specific, so they may not always apply to general radiology use cases.
In the MI-CDM paper (Park et al.), the proposed approach was:
- RadLex and LOINC for radiological findings and measurements
- SNOMED CT for anatomical location
RadLex offers rich hierarchies for imaging findings, for example:
Clinical finding → Pathophysiologic finding → Mechanical disorder → Fluid disorder → Effusion → Pleural effusion (RID34539).
Another potential resource is RadLex Common Data Elements (CDEs), which define standardized “key–value” pairs for structuring radiology reports. They are not yet OMOP standard concepts, but could be relevant for your use case.
The OHDSI Medical Imaging Workgroup (WG) is actively discussing how to incorporate such imaging vocabularies into OMOP, and your scenario could contribute meaningfully to that conversation. I’d encourage you to join a meeting so we can explore it together.
3. Anatomical Location Representation
For anatomical regions imaged, the MI-CDM image_occurrence.anatomic_site_concept_id should follow the “lowest level of granularity” from DICOM’s Defined Terms or Anatomical Region codes (often drawn from SNOMED CT or LOINC).
The DICOM2OMOP paper has mapped these DICOM anatomical terms into OMOP vocabularies, with the corresponding lists available on GitHub. In practice, I interpret “lowest level of granularity” as the most specific term available, but this might be worth confirming with the WG as interpretations could vary.
For the vocabulary and note-related topics, it would be great to continue the discussion in the WG. Meetings are biweekly on Wednesdays at 7 AM and 7 PM ET, with the next meeting scheduled for Wednesday, September 3 at 7 AM ET. I hope to see you there.