We have submitted (and funded) an enhancement request to enable use of datetime in cohort definitions. The intent of this forum post is to facilitate community discussion about this topic. Although this feature was added as a milestone for Atlas 2.15, we’ve heard recent requests for more discussion to ensure that this would not break existing functionality.
The details of the need and request are here. That link includes discussions about details of implementation options. Below is an excerpt of the enhancement request.
Problem Statement
Multiple US electronic Clinical Quality Measure (eCQM) require datetime calculations when doing computations from electronic health records. Public domain examples are published here. The trend for needing datetime logic appears to be increasing.
In many cases, it is possible to create versions of those eCQM as Atlas cohorts, and then adapt the SQL to use datetime calculations instead of datetime. Although this may work for single institutions, it does not lend itself to developing these as phenotypes that can be run in a network study.
Examples:
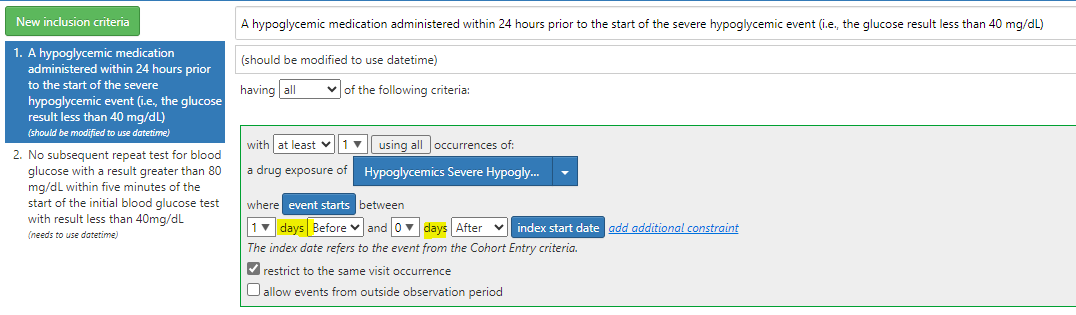
- Hospital Harm - Severe Hyperglycemia. Checks for days when very high glucose measurements are present, but based upon rolling 24 hour periods starting from the time of ED or hospital admission (instead of calendar dates). Thus, the index date is actually a datetime.
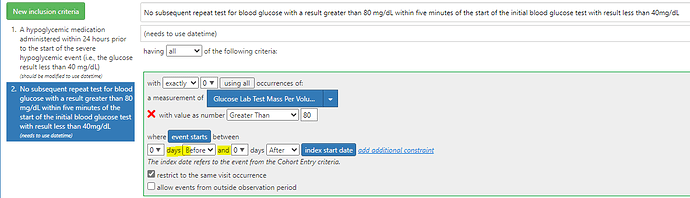
- Hospital Harm - Severe Hypoglycemia. Checks whether follow-up glucose measurements occurred within 5 minutes after critically low measurements.
- Hospital Harm - Opiate-Related Adverse Events. Checks whether opioid antagonists had to be administered within 12 hours after administration of an opioid.
Current Behavior
The Atlas GUI for selecting timespans (e.g. between two dates) only supports whole-number date logic.
Desired behavior
Augment the Atlas GUI to allow for whole number datetime interval logic, with options to specify “minutes”, “hours”, or “days” instead of only “days”.
For simplicity to the users, the internal logic should know to use datetime interval logic instead of date logic whenever “minutes”, or “hours” are selected. This would eliminate the need for cohort authors to choose between “index start date” and “index start datetime”.
For example, if you want to know that an event occurred within 24 hours of admission, you would use an interval of 24 hours (which would use datetime logic) rather than 1 day (which would use date logic).
Here is example where I’d want to use 24 hours:
And here is example where I’d want to use minutes:
Feature Request Scope
- Make the word “days” a drop-down box like “Before/After” than has choices {days, hours, minutes)
- Augment the JSON to support this. I presume you want backwards compatibility. So, perhaps keep the existing syntax, but modify it to add optional “TimeUnit” and “TimeUnitValue” fields, where “TimeUnit” can have values “Days”, “Hours”, “Minutes”; and “TimeUnitValue” would have the desired value (that is currently show in the “Days” field? Below is example of how such time windows are currently represented in JSON:
"StartWindow": {
"Start": {
"Days": 9,
"Coeff": -1
},
"End": {
"Days": 1,
"Coeff": -1
},
- Augment the generated SQL to use time interval logic whenever a TimeUnit other than “Days” is selected. Retain the current logic when “Days” is selected. Note. I have opened a feature enhancement issue with SqlRender to see if are able to generate interval time translations across all supported databases. Per GPT-4, the translations looks pretty straight forward.
Note, although not all data source have datetime-level data, the 5.3 OMOP data model and above all require datetime fields. By convention, those are populated with the same value that are in the matching data field. So, if this enhancement is added, phenotypes using seconds/minutes/hours should work (without breaking) on any 5.3 and above OMOP datasets. Data contributors would need to clarify which subset (if any) of their data has true datetime values, and Researchers would need to take that into consideration.
It would be possible to augment Data Quality Dashboard to test which data sources have actual datetime values, plus internal consistency between those datetime values and the regular date values.