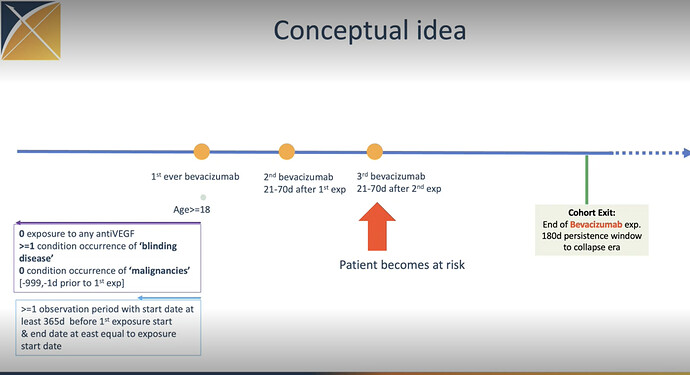
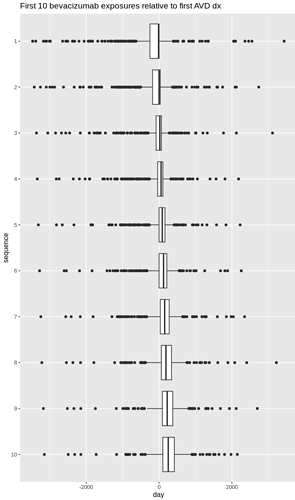
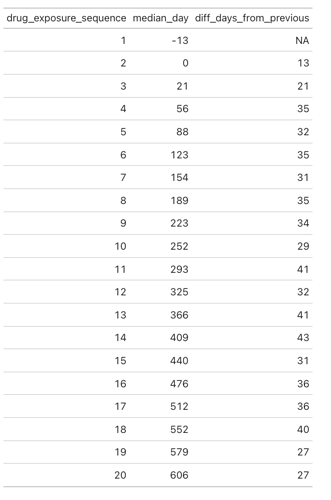
I tried to create a simple test of this cohort definition to test if it is indeed possible to pick up persons with only three bevacizumab exposures. However I could not create a passing test with Eunomia. I’m not sure what I’m doing wrong but here is my code. In addition to the actual cohort I also tried a very simple bevacizumab cohort definition.
library(dplyr, warn.conflicts = F)
# devtools::install_github("OHDSI/Eunomia", "develop")
connectionDetails <- DatabaseConnector::createConnectionDetails(
dbms = "duckdb",
server = Eunomia::eunomiaDir("GiBleed", dbms = "duckdb")
)
cohortDefinitionSet <- ROhdsiWebApi::exportCohortDefinitionSet(
baseUrl = "https://atlas-demo.ohdsi.org/WebAPI",
cohortIds = c(1782491, 1782502)
)
#> Fetching cohortId: 1782491
#> Fetching cohortId: 1782502
nm <- CohortGenerator::getCohortTableNames()
con <- DatabaseConnector::connect(connectionDetails)
#> Connecting using DuckDB driver
CohortGenerator::createCohortTables(connection = con,
cohortDatabaseSchema = "main",
cohortTableNames = nm)
#> Currently in a tryCatch or withCallingHandlers block, so unable to add global calling handlers. ParallelLogger will not capture R messages, errors, and warnings, only explicit calls to ParallelLogger. (This message will not be shown again this R session)
#> Creating cohort tables took 0.24secs
CohortGenerator::generateCohortSet(connection = con,
cdmDatabaseSchema = "main",
cohortDatabaseSchema = "main",
cohortDefinitionSet = cohortDefinitionSet,
cohortTableNames = nm)
#> 1/2- Generating cohort: [SOS Phenotype Devt] bevacizumab exposures cohort SHELL
#> Executing SQL took 0.481 secs
#> 2/2- Generating cohort: bevacizumab exposure
#> Executing SQL took 0.0326 secs
#> Generating cohort set took 1.05 secs
DatabaseConnector::querySql(con, "select * from main.cohort")
#> [1] COHORT_DEFINITION_ID SUBJECT_ID COHORT_START_DATE
#> [4] COHORT_END_DATE
#> <0 rows> (or 0-length row.names)
# looks like the image of this definition on GiBleed Eunomia is empty
# Let's now insert records for a person, say person Id 1
DBI::dbExecute(con, "delete from main.condition_occurrence where person_id = 1")
#> | | | 0% | |======================================================================| 100%
#> Executing SQL took 0.00665 secs
#> numeric(0)
DBI::dbExecute(con, "delete from main.drug_exposure where person_id = 1")
#> | | | 0% | |======================================================================| 100%
#> Executing SQL took 0.00811 secs
#> numeric(0)
drugs_to_add <- tribble(
~drug_exposure_id, ~person_id, ~drug_concept_id, ~drug_exposure_start_date, ~drug_exposure_end_date,
100000, 1, 1397141, "2000-01-01", "2000-01-02",
100001, 1, 1397141, "2000-02-01", "2000-02-02",
100002, 1, 1397141, "2000-03-01", "2000-03-02"
) %>%
mutate(drug_type_concept_id = 32817)
conditions_to_add <- tibble(
condition_occurrence_id = 1000000,
person_id = 1,
condition_concept_id = 374028,
condition_start_date = "1999-12-01",
condition_end_date = "1999-12-02",
condition_type_concept_id = 32020)
DBI::dbWriteTable(con, "drug_exposure", drugs_to_add, append = TRUE)
#> Inserting data took 0.018 secs
DBI::dbWriteTable(con, "condition_occurrence", conditions_to_add, append = TRUE)
#> Inserting data took 0.0254 secs
# check that these records are in the cdm now
DBI::dbGetQuery(con, "select * from main.drug_exposure where person_id = 1")[,1:4]
#> drug_exposure_id person_id drug_concept_id drug_exposure_start_date
#> 1 100000 1 1397141 2000-01-01
#> 2 100001 1 1397141 2000-02-01
#> 3 100002 1 1397141 2000-03-01
DBI::dbGetQuery(con, "select * from main.condition_occurrence where person_id = 1")[,1:4]
#> condition_occurrence_id person_id condition_concept_id condition_start_date
#> 1 1e+06 1 374028 1999-12-01
DBI::dbGetQuery(con, "select * from main.observation_period where person_id = 1")[,1:4]
#> observation_period_id person_id observation_period_start_date
#> 1 1 1 1949-01-28
#> observation_period_end_date
#> 1 2019-05-24
# So we should have at least one person who meets the cohort definition right?
CohortGenerator::generateCohortSet(connection = con,
cdmDatabaseSchema = "main",
cohortDatabaseSchema = "main",
cohortDefinitionSet = cohortDefinitionSet,
cohortTableNames = nm)
#> 1/2- Generating cohort: [SOS Phenotype Devt] bevacizumab exposures cohort SHELL
#> Executing SQL took 0.468 secs
#> 2/2- Generating cohort: bevacizumab exposure
#> Executing SQL took 0.0306 secs
#> Generating cohort set took 0.78 secs
DatabaseConnector::querySql(con, "select * from main.cohort")
#> [1] COHORT_DEFINITION_ID SUBJECT_ID COHORT_START_DATE
#> [4] COHORT_END_DATE
#> <0 rows> (or 0-length row.names)
# But still nothing....
# Lets try adding a fourth drug exposure
drugs_to_add2 <- tribble(
~drug_exposure_id, ~person_id, ~drug_concept_id, ~drug_exposure_start_date, ~drug_exposure_end_date,
100003, 1, 1397141, "2000-04-01", "2000-04-02"
)
DBI::dbWriteTable(con, "drug_exposure", drugs_to_add2, append = TRUE)
#> Inserting data took 0.0104 secs
DBI::dbGetQuery(con, "select * from main.drug_exposure where person_id = 1")[,1:4]
#> drug_exposure_id person_id drug_concept_id drug_exposure_start_date
#> 1 100000 1 1397141 2000-01-01
#> 2 100001 1 1397141 2000-02-01
#> 3 100002 1 1397141 2000-03-01
#> 4 100003 1 1397141 2000-04-01
DBI::dbGetQuery(con, "select * from main.condition_occurrence where person_id = 1")[,1:4]
#> condition_occurrence_id person_id condition_concept_id condition_start_date
#> 1 1e+06 1 374028 1999-12-01
CohortGenerator::generateCohortSet(connection = con,
cdmDatabaseSchema = "main",
cohortDatabaseSchema = "main",
cohortDefinitionSet = cohortDefinitionSet,
cohortTableNames = nm)
#> 1/2- Generating cohort: [SOS Phenotype Devt] bevacizumab exposures cohort SHELL
#> Executing SQL took 0.464 secs
#> 2/2- Generating cohort: bevacizumab exposure
#> Executing SQL took 0.0284 secs
#> Generating cohort set took 0.72 secs
DatabaseConnector::querySql(con, "select * from main.cohort")
#> [1] COHORT_DEFINITION_ID SUBJECT_ID COHORT_START_DATE
#> [4] COHORT_END_DATE
#> <0 rows> (or 0-length row.names)
# still nothing
DatabaseConnector::disconnect(con)
Created on 2023-04-12 with reprex v2.0.2
Update: I think the issue is that the concepts being used are not in the mini-eunomia vocabulary. I tried adding them but that gave me a different error when I generated the cohorts.