SARS-CoV-2, the virus that causes COVID-19, change over time. Different variants have significant impact on virus’s properties, such as how easily it spreads, the associated disease severity, or the performance of vaccines, therapeutic medicines, diagnostic. World Health Organization (WHO) currently designated four Variants of Concern. My question is: where are the SARS-CoV-2 variants in OHDSI vocabulary?
Hi @QI_omop,
There are a couple of LOINC codes:
- SARS-CoV-2 (COVID-19) variant [Type] in Specimen by Sequencing
- SARS-CoV-2 (COVID-19) clade [Type] in Specimen by Molecular genetics method.
- SARS-CoV-2 (COVID-19) lineage [Identifier] in Specimen by Molecular genetics method
- SARS-CoV-2 (COVID-19) variant interpretation in Specimen Narrative.
Unfortunately, we’re missing LOINC answers for 1-3 because the concepts were added in LOINC 2.70 version, but we’re still on 2.69. Within the next LOINC release in OMOP, it will be all set.
BTW, the variant lists are conflicting a bit. LONC doesn’t provide the delta variant, while Nigerian (B.1.1.238) and Cluster 5 are included.
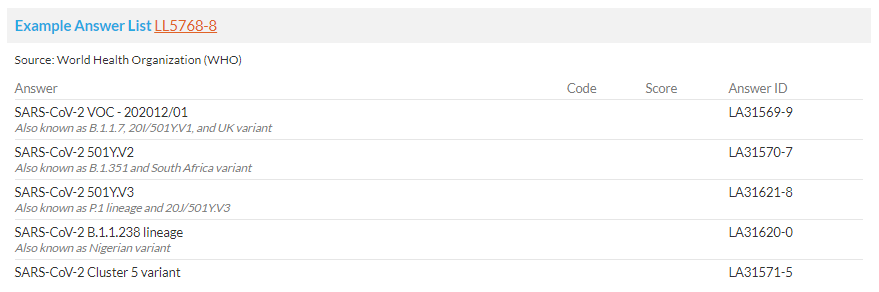
Note:
The source for these outcomes is:
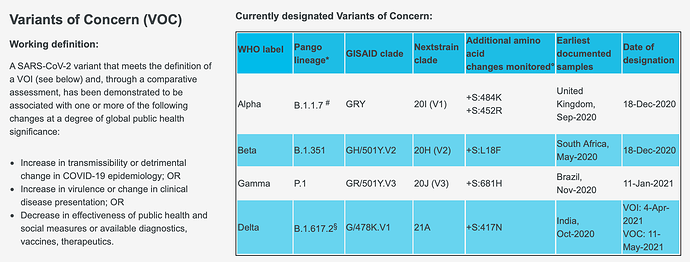
Breaking down these outcomes from the WHO COVID-19 Core CRF, here are the VOCs:
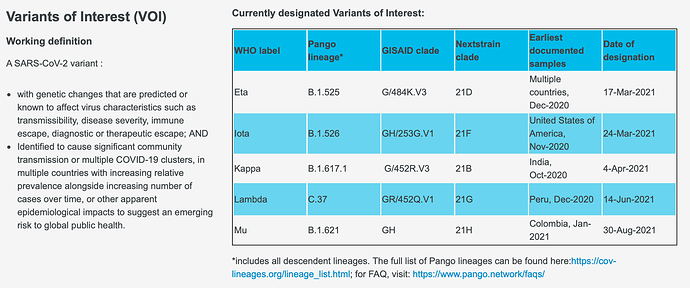
And here are the VOIs:
LOINC 2.71 doesn’t capture these VOIs either by lineage or clade. At the same time the 05Jul21 version of the WHO COVID-19 Core CRF is in the process of being rolled out in parts of Sub-Saharan Africa. So there is a vocabulary shortfall that maybe doesn’t spell well for genomic surveillance.
A team I work on is just now completing a beta version of an OMOP implementation guide for the WHO COVID-19 Core CRF. What concepts should we use in the Outcomes section for the VOIs that the WHO has included?
Hi @JayGee ,
LOINC is not quite keeping up with the pace new variants are described. But I cannot blame them, we too would need some time to adopt their concepts once they are available. The delta variant is now included in the LOINC 2.71 version. It is a bummer that you cannot always put the WHO label in the value of the measurement (as it is numeric only) and only add the LOINC answer concept in value_as_concept_id if you really have it already in the OMOP LOINC vocabulary. Not a great alternative is to fall back to observation as we would have a mixed approach then. @Alexdavv , do you have a good idea how to approach this?
I wouldn’t go with linage or clade concepts unless the source explicitly says this.
I think the best option is mentioned
…while narrative one concept just creates an ambiguity.
As far as LOINC is missing the specific types (answers) to be used as value_as_concept_id, I would simply recommend the Greek letters:
Would you recommend SARS-CoV-2 (COVID-19) variant [Type] in Specimen by Sequencing together with the SNOMED Greek letters as value_as_concept_id as a best practice in federated studies in SSA that ETL REDCap and other databases that perform WHO COVID-19 Core CRF capture into OMOP? @msuchard, @Andy_Kanter, @chifundok and @Andrew, is this something we might talk about at our next OHDSI Africa Chapter Monthly Meeting?
Right.
Yes, but only for those that are not added to OMOP as LOINC answers as the moment of ETLing/concept set building.
LOINC already has 6 variants, but missing others.
As for now, we’re missing all of them in OMOP. But we’ll be gradually adding LOINC answers and, hopefully, LOINC authors will add all missing at some point.
You’re not interested in capturing the types based on other classification systems (Pango, GISAID, Nextstrain), right?
Using the Greek letters with a gradual replacement to proper LOINC answers sounds reasonable. Once LOINC answers are added to the OMOP vocabulary, you can adjust the mappings and concept sets even keeping the Greek letters (together with proper LOINC answers) in order to support both vocabulary versions.
The only potential issue that comes to my mind is the Sequencing is not the only method that can be used for types detection. As for European guidelines, PCR/antigen assays can be used in screening, while sequencing is the only confirmatory technique. The CDC’s guidelines doesn’t mention other methods, but I would double check with the NS3 team.
If the source data specify the method and we want to distinguish it in OMOP, the option would be the local OMOP Extension concepts (before LOINC has a better representation).
Thank you.
By the way @Alexdavv the solution is a little like a rube goldberg machine and I like it (not that that matters much)
Sequencing may not be an issue. Perhaps in the middle of the July2021 WHO COVID-19 Core CRF which covers “at any time during the visit occurrence”, the ask is for the results of PCR testing. LOINC handles this at various levels of specificity. However, it isn’t until the Outcomes section that there is an ask for variants in connection with a diagnosis based on laboratory results. That would be the solution you are proposing.
There is a second use case we also cover in the implementation guide. This is a sentinel surveillance use case where a sample is collected in the field and sent directly to a lab for sequencing. This use case is happening with more frequency now because the Gates Foundation has begun to build/fund genomic surveillance systems in LMICs. Your solution also handles this use case.
Maybe more discussion is needed about phasing out the Greek letters since the WHO intended them as an alternative to nomenclatures that identify countries. That discussion is at another level…
In any event I will build one or two measurement records for each use case and post them here along with rows from the implementation guide that direct the record creation.
Looks like the topic gets more and more complicated.
The variant vocabulary already exists in OMOP (SNOMED Organism). But it only contains Variants of Concern (VOC) shown below:
- Genus Alphacoronavirus 44783751
- Genus Betacoronavirus 44783752
- Genus Deltacoronavirus 44783753
- Genus Gammacoronavirus 44783754
We need Variants of Interest (VOI) to be added as OMOP extension, such as Delta Plus, Lambda, Mu etc.
See this detailed taxonomy of the Orthocoronavirinae family:
This is another stuff much more higher in the taxonomy.
SARS‑CoV‑2 is defined as Virus
of Severe acute respiratory syndrome–related coronavirus species (missing in SNOMED)
of Sarbecovirus subgenus
of Betacoronavirus Genus.
The latter is the one you’ve mentioned.
In fact, SNOMED has nothing below the SARV-COV-2 virus - neither variants nor types.
Haha.
This is pretty much how vocabs complement each others.