Folks,
Thank you kindly for the opportunity to present at the community call this past Tuesday. I’ll be re-reviewing your questions, concerns, and suggestions from the recording.
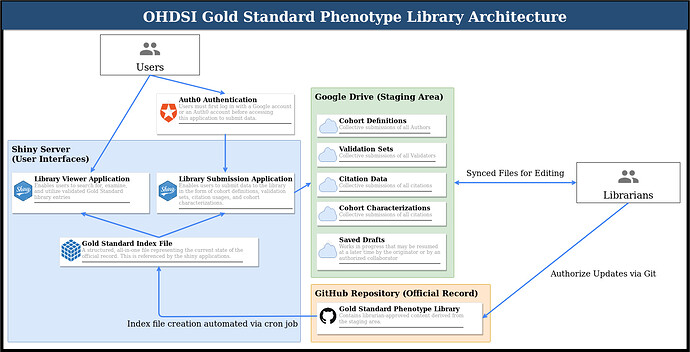
In advance of the software demonstration at the symposium, I wanted to release this updated architecture diagram, which I believe more clearly articulates the cycle being proposed. After the symposium, I’ll be writing up the technical specifications in more detail, but the diagram below outlines the process. I expect to have a working proof of concept for this particular configuration at the demo.
To walk through this:
- Users have two Shiny applications which they can use to interface with the library in different ways:
-
The viewer application for read-only activities (i.e. examining, searching/filtering, and/or downloading cohort definitions). This is a space where we can display whatever kinds of charts, diagrams, tables, etc. people find useful when they are trying to locate a cohort definition.
-
The submission application to propose adding new data to the library, which requires authentication with a Google account. Currently, this is being carried out with Auth0 using the auth0 R package. The idea here is that the librarians would have access to the full Auth0 dashboard, which has a lot options that comes with it (details left to future tech specs document).
-
The types of data being submitted I’ve been calling “modes of submission”. You might want to submit a cohort definition of your own, or submit validation data for someone else’s definition. You might want to submit a citation for a library definition that’s been used in a publication, or you might want to submit a cohort charaterization so others can anticipate what their cohorts may look like when they execute the cohort definition at their site.
-
Whatever the case, when data are submitted using the corresponding form within the Library Submission Appplication, the data are processed and then uploaded to a Google Drive service account dedicated to the library.
-
Librarians can then clone the gold standard phenotype library repository and sync to the Drive folder. Every librarian who does this becomes one with the staging area, so to speak. Any changes made online on Drive or locally on the librarian’s computer will show up for all other librarians as the changes sync. This is also true as the Shiny application uploads its submission data to Drive; the newly uploaded data will sync for all librarians.
-
The staging area is not the official record though, as proposed submissions may take varying degrees of consideration (peer review) before they are ready to be elevated to OHDSI’s official library. When a staged change is ready to become official, the librarians can add it to the repository via standard git commands, thereby taking ownership of that particular update.
-
Before the content can reach back to the viewer/submission applications, an index file is necessary to compile the data into a single object. This essentially allows the applications to run and load far faster than they would if they had to query the repository and perform all of the calculations at runtime. A simple example of this is with metrics. Recall that we ask for raw true/false positive/negative values; we therefore calculate the sensitivity, specificity, PPV, NPV, F1 score, and accuracy “behind the scenes” so that the applications can display the numbers without first having to calculate them. Another example is provenance, where we calculate connected components so that we can display a network graph for a given cohort definition to show all of its relationships to other definitions. The index file is also an opportunity to insert other advanced calculations and pre-loading mechanisms that have not yet been foreseen.
-
Finally, a cron job can automatically rebuild the index file on the server by periodically checking the index file timestamp against the timestamp of the latest commit to the official record. If something has been pushed to the official record after the index file was last made, then the index is due for an update and will be rebuilt with the updated data from the official record. This advantages the librarians, because once they have pushed an update to the official record, they don’t need to worry about anything else, as the index will automatically update on its own. We could also consider a git hook to tie the automation more precisely to a commit event, but that would place more burden on the librarians to ensure the hook is configured and executing properly.
-
This completes the cycle! When a user checks back into the library applications, the updates that the librarians have brought to the official record will appear.
As a mentioned in the call, the requirements of the library are conceptually separate from the technical specifications, but I think both can advance simultaneously to some extent. A good example of this was when @Patrick_Ryan raised the idea that having cohort characterizations would be a good mode of submission. From a requirements perspective, this currently needs its own set of data elements to be specified; precisely, what do we expect users to submit when they add their own cohort characterization? However, from a technical perspective, it’s relatively easy to extend the Library Submission Application to incorporate one more type of submission.
I’ll be on the line during our regularly scheduled WG meeting (Tuesdays @ 10am ET) for those who wish to discuss this further. The week after, we’ll cancel the meeting since most of us will be at the symposium, which I’m very much looking forward to! ![]()
Cheers,
Aaron