Hello Everyone,
I require inputs from the data analysts here on how can I tackle/address the below scenario?
I have below tables in my database with record count as shown below
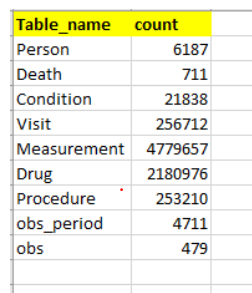
I would like to join all these tables and make one final/main table. It’s like X1,X2,X3…Xn are the inputs and Y will be the output label. I know there will be lot of NA’s but that’s fine
I tried the below approaches but nothing helped
-
Currently we use PostgreSQL db and the join query runs for a long time and doesn’t provide any output at all even after the joining keys are indexed.
-
I tried joining using Python pandas but didn’t help either. This also keeps running but no output as yet
-
I thought it would be better to summarize the
measurementtable to reduce the rows count (but increase the column count). Due to database limitation of 1664 columns, this doesn’t help either because our summarized output will have more than 1664 columns. -
I also thought of parallel processing but this one doesn’t help either. I tried with 4 core and 24 core cpu. It was running but didn’t produce any output.
None of the above approaches produced any output. At least not within an hour. But I would like to have this final table in 5 mins time? Is it possible? Can pyspark help? I don’t want to use python Dask(I see this often compared with spark) as it doesn’t have the required functions like pandas unstack.
Any suggestions/easy way to merge/join all these tables into one? If you think Measurement table is causing the issue, the only option I am left with is to sample (stratify) the data?
