Hello. I ask some question of covariates.
I’m struggling with using CohortMethod pacakges in R and when I run getDbCohortMethodData() function, foutunately it runs.
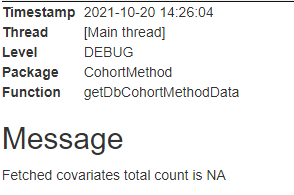
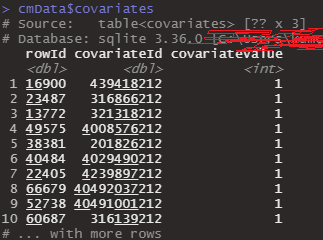
But the point is the result of that function show that it has no covariates like below.
Covariates:
Number of covariates: NA
Number of non-zero covariate values: NA
Before I run getDbCohortMethodData(), I set covariates with run createDefaultCovariateSettings() and when I check the concept ID of covariates that I will exclude it shows it.
I don’t know what is the problems and how to solve this one. Please help me