Hi, I am trying to implement PheValuator on my OMOP CDM data. I have seven different diseases of interest, so I am trying to find a general formula for PheValuator cohort definitions that works for all. For each disease I identify a key OMOP code that represents the disease (i.e. 201826 for T2D) though all diseases of interest are difficult to identify in the EHR and care needs to be taken in their phenotype definition.
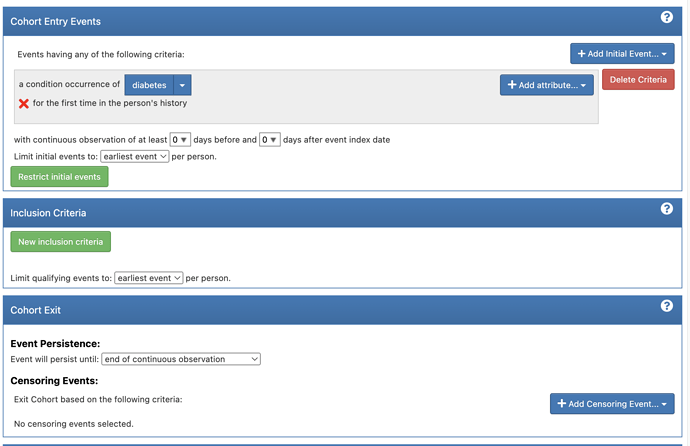
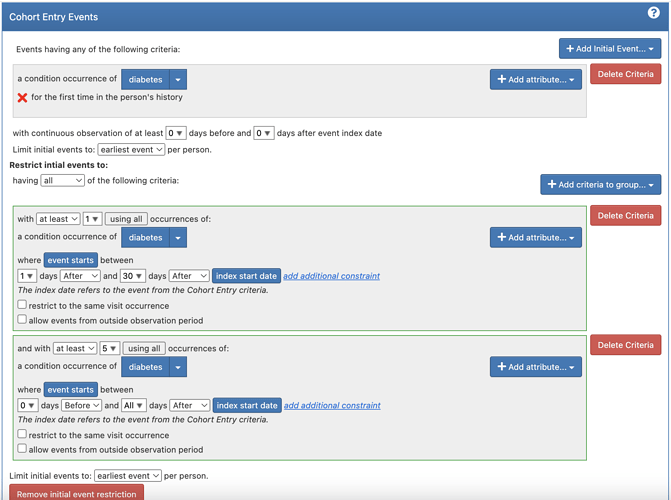
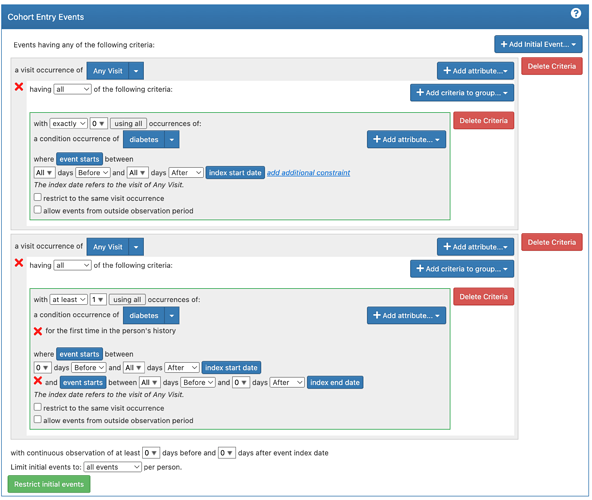
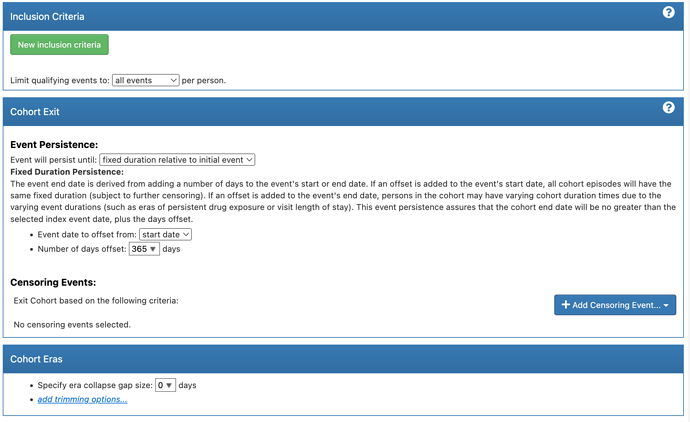
After looking through the paper and vignette I have come up with the following definitions for my cohorts (note the “diabetes” concept set is the key OMOP code + descendants)
-
xSens: ( = Prevalence)
-
xSpec:
(here I alter 30 days after to 60 days after for rare diseases)
- Evaluation:
Are these definitions adequate? One concern I have is from a sentence in the Discussion of the PheValuator paper which says: “This approach is not applicable in situations where there is only a single concept that is used to define a disease as the xSpec PA and the predictive model would both require use of that concept, which would introduce circular logic.” - is this meant to i) indicate more broadly that only complex disease like T2D should be studied using PheValuator or ii) describe a scenario where one cannot leverage one key code + descendants across all definitions?
Thanks!