I very much like what @jswerdel showed us here. The evaluation clearly illustrates that PPV alone is not enough to evaluate the phenotype. And as @agolozar points out, there is a need to better understand the performance of the phenotypes across all participating data partners. Now, we can’t really expect all of them to do an extensive chart review nor it is feasible to do it on claims data sources. We can use Phevaluator to estimate the performance but it would be really nice to also get an idea of what type of patients we include, their disease history, etc. - what @david_vizcaya calls a narrative/clinical validity.
Since patient profiles (or other patient descriptions) enable us to examine such clinical validity and incorporate our observations into a better version of the phenotype. For example, as @Gowtham_Rao said, we can learn from false-positives and refine the definitions. I’m thinking we can operationalize some of the criteria that make us think that the case is false positive, like:
- Presence of alternative diagnosis after the index date. It is quite possible that we may observe alternative diagnoses before the index date (especially for complex conditions) but by that time we expect them to be ruled out. Hard problem here if to distinguish between possibly co-occurring disorders and mutually exclusive disorders;
- Implausible data density/visit context. For example, if multiple myeloma requires impatient stays, having 0 events for a year after the index date is suspicious;
- Absence of treatment and/or diagnostic procedures. Tricky, because patients can legitimately be diagnosed outside of the system. Also, sometimes they don’t get specific treatment - CKD is a good example.
- Implausible age+gender. For example, in one of OHDSI studies we even incorporated age in our asthma drug-based definition.
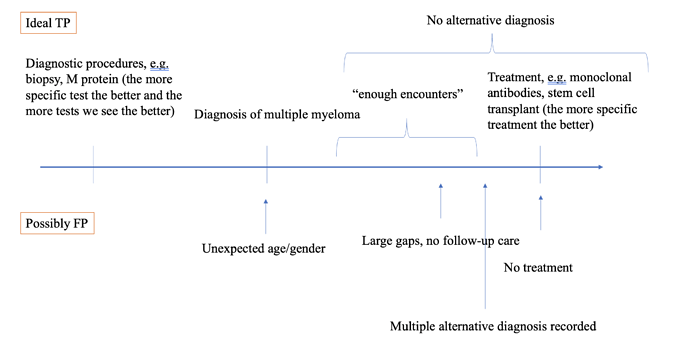
In a very simplified form, we would expect true positives and true negatives to look something like this:
And in a real cohort, we can observe a whole spectrum of patients from “very likely true positive” to “very unlikely true positive”. What I struggle with here is how to come up with rules/examples for false negatives - those patients who do not have the necessary elements of the phenotype in their data yet have the disease (and how to figure out they have the disease).