Phenotype Phebruary Day 23 – Hidradenitis Suppurativa
Today is the 23rd of Phebruary and I will be sharing findings from the Hidradenitis Suppurativa (HS) Phenotype. I collaborated with @JoelSwerdel to develop this immunology phenotype. To be clear we did not have a specific use case for application of the HS phenotype.
Since I suspect many collaborators may not know what this phenotype entails, I’ll start with a clinical overview.
Hidradenitis suppurativa (HS) is a chronic, recurring, inflammatory disease of the skin. Clinically, subjects have nodules, draining skin tunnels (sinus tracts), abscesses, and bands of severe scar formation in the intertriginous skin areas such as axillary, groin, perianal, perineal, and inframammary regions (Alikhan A et al). HS typically begins in the second or third decade of life (Alikhan A et al; Liy_Wong C et al). Patients with HS suffer from metabolic, psychiatric, and autoimmune disorders (Tiri H et al). Treatment is based on severity of symptoms with wound care, pain management, topical or oral antibiotics, debridement as needed.
Following my fellow phenotypers, after we understood the clinical description, we worked with our internal expert Ms. Gayle Murray who generated a systematic literature review for HS. The literature review used the following terms:
(“hidradenitis suppurativa”[MeSH Terms] OR (“hidradenitis”[All Fields] AND “suppurativa”[All Fields]) OR “hidradenitis suppurativa”[All Fields])
AND
(“retrospective cohort”[All fields] OR (“Epidemiology”[mh]) OR “Epidemiologic Methods”[mh] OR (phenotype[TIAB]) OR (insurance OR claims))
AND
(((((((((((((((((((((Medicaid) OR (medicare)) OR (truven)) OR (Optum)) OR (Medstat)) OR (Nationwide Inpatient Sample)) OR (National Inpatient Sample)) OR (PharMetrics)) OR (PHARMO)) OR (Validation study[Publication Type])) OR (positive predictive value)) OR (ICD-9[Title/Abstract])) OR (ICD-10[Title/Abstract])) OR (claims[Title/Abstract])) OR (insurance[Title/Abstract])) OR (General Practice Research Database)) OR (Clinical Practice Research Datalink)) OR (IMS[Title/Abstract])) OR (patient registry)) OR (electronic medical records[Text Word])) OR (electronic health record[Text Word])) OR (((“Databases, Factual”[Mesh]) OR “Denmark/epidemiology”[Mesh]) OR “United States Department of Veterans Affairs/organization and administration”[Mesh])
NOT
((“Clinical Trial”[pt] OR “Editorial”[pt] OR “Letter”[pt] OR “Randomized Controlled Trial”[pt] OR “Clinical Trial, Phase I”[pt] OR “Clinical Trial, Phase II”[pt] OR “Clinical Trial, Phase III”[pt] OR “Clinical Trial, Phase IV”[pt] OR “Comment”[pt] OR “Controlled Clinical Trial”[pt] OR “Letter”[pt] OR “Case Reports”[pt] OR “Clinical Trials as Topic”[Mesh] OR “double-blind”[All] OR “placebo-controlled”[All] OR “pilot study”[All] OR “pilot projects”[Mesh] OR “Prospective Studies”[Mesh])) OR (“Genetics”[Mesh])
After excluding many unrelated articles, the review identified 30 relevant articles and, of those, five had validation metrics (Kim et al; Shlyankevich et al; Garg et al 2017; Garg et al 2017; Strunk 2017). Note that these studies used chart review to determine diagnostic accuracy measures. Of those five studies there were three that used large insurance claims databases and the remaining two studies used a hospital-based database. We generated the cohorts for one study to show how the authors’ study results compared to ours using our data and rigorous methodical approach. We will discuss this comparison later in this post.
After we reviewed the literature and determined the relevant codes being used we converted these ICD codes to standard SNOMED concepts using ATLAS. We then utilized PHOEBE to help us understand if we missed any relevant codes and used the two approaches to develop the concept set, which was:
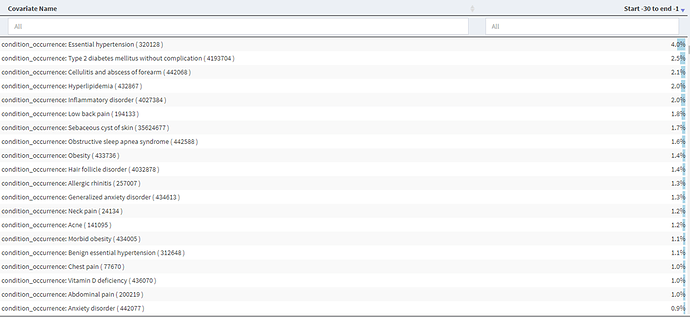
We then began building our cohort definitions. We used the Cohort Diagnostics tool (thank you @Gowtham_Rao) to examine the conditions and drugs in the time prior to an initial diagnosis of HS to see if there was evidence of index date misclassification and found no evidence as there is an absence of high proportion covariates (>5%) in the -30 to -1 days prior to index (CCAE results below and MDCD very similar). BTW – here is the link to the CohortDiagnostics results (NOTE: search for hidradenitis to see the cohorts and associated information)
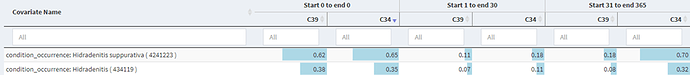
So with that in mind we then investigated the use of 1 code versus 2 codes to get a sense of the impact on the specificity of the algorithm. For a single condition code, we saw in Cohort Diagnostics the following.
This shows a large drop off in the proportion of those with an HS code in the time after index, so we developed a more specific definition requiring a second diagnosis code in the 31 – 365 day after index and found:
Which showed a modest increase in the in the proportion of those with an HS code in the time after index. The measure we looked at was in the 1-30 day post index as the 31-365 day measure included required codes in cohort C34.
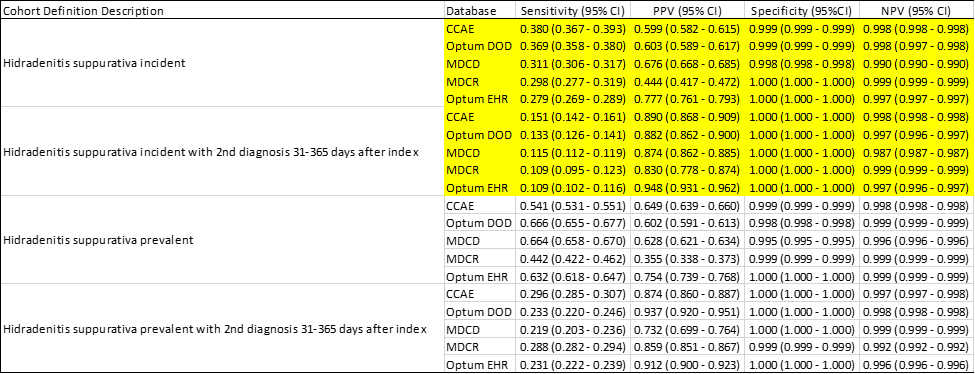
We ran PheValuator (thanks @JoelSwerdel) on the cohorts and found:
We see a large increase in positive predictive value (PPV) when we compared a single code algorithm to an algorithm requiring a second code. This, though, came at a cost in sensitivity. We also compared our results for the Kim et al study. Kim et al used data from the Massachusetts General Hospital database and reported an increase in PPV with an increasing number of HS diagnostic codes (2 codes: 81% vs 5 codes: 97% etc). Our study replicated Kim et al cohorts and found an increase in PPV with use of five or more diagnostic codes compared to at least two HS diagnostic codes (5+ code mean: 84% vs 2 code mean: 59%) similar to, albeit lower than, the published results.
Recall from the outset of this post that there was no use case for this phenotype but rather our goal was to populate our phenotype library with HS phenotypes for future use. In our final analysis, we concluded that we should make four algorithms available for use with different “fit for function” purposes. If your research requires a high sensitivity algorithm, possibly looking at HS where you don’t want to miss cases, then you might consider cohort definition “Hidradenitis suppurativa prevalent”. If you want to ensure that you have prevalent cases of HS with a high probability of truly having the condition, for instance in drug treatment pathway studies where you want only those subjects with HS, you may want to use “ Hidradenitis suppurativa prevalent with 2nd diagnosis 31-365 days after index”. If your research requires incident cases of HS, then you may want to use either “ Hidradenitis suppurativa incident” if you want to maximize sensitivity or “ Hidradenitis suppurativa incident with 2nd diagnosis 31-365 days after index” if maximizing specificity is your requirement.