Team:
Today, while many in the US are preparing for their Super Bowl football/commercial viewing parties, I’d like to discuss phenotyping Attention Deficit Hyperactivity Disorder (ADHD). This is work that several colleagues (@ericaVoss @jweave17 @Jill_Hardin @conovermitch ) and I conducted over the past couple years that yielded a bunch of lessons learned the hard way. But the experience gave us insights to some complementary recommended practices that I’ve carried forward in my own research, so I thought I’d walk through the development process to show the pitfalls and ways to overcome them.
Clinical description:
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder characterized by patterns of inattention or impulsive behaviors that interferes with functions. ADHD can be classified by the most common symptoms, be it ‘predominantly inattentive’ presentation- where patients may struggle to organize or complete tasks, follow directions, or remember details of daily activities- or ‘predominantly hyperactive-impulsive’ presentation - where patients are constantly moving around or talking or interrupting when not appropriate. ADHD is most commonly diagnosed in childhood (though can occur with onset in adults), through physician examination and evaluation of ADHD symptoms over time and its impact in social settings (and ruling out alternative diagnosis from other mental health disorders or environmental factors). Management of ADHD may involve behavioral therapy or pharmacologic treatment (including stimulants such as methylphenidate, amphetamine, lisdexamfetamine, and non-stimulants such as atomoxetine and guanfacine).
Phenotype development:
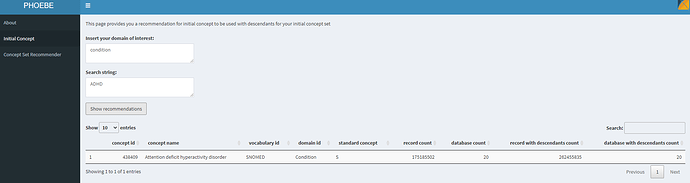
Let’s follow some of the practices we’ve discussed on prior threads. We’ll start with PHOEBE to find our starting point in the OHDSI vocabulary:
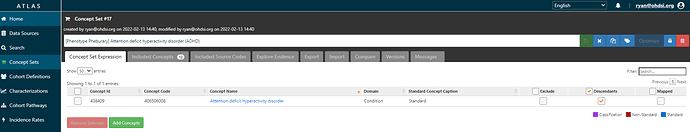
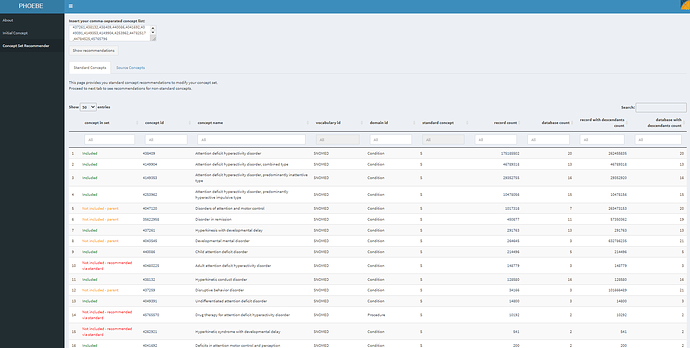
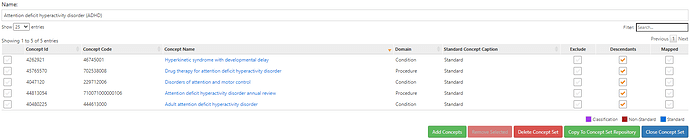
Ok, makes sense, ‘Attention deficit hyperactivity disorder’, a standard concept in SNOMED, sounds reasonable to begin with. We see that 20 databases have records with this concept, and it looks like there are many records for descendants of this concept, so probably picking ADHD + descendants would be useful to feed into PHOEBE to see if we can any other recommendations. I’ll use ATLAS to create a conceptset expression that we can build up:
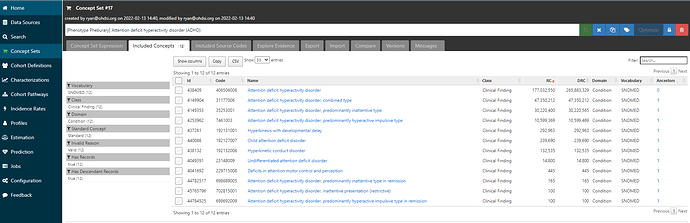
We see there are 12 included concepts, many of which have substantial record counts and seem reasonable to include:
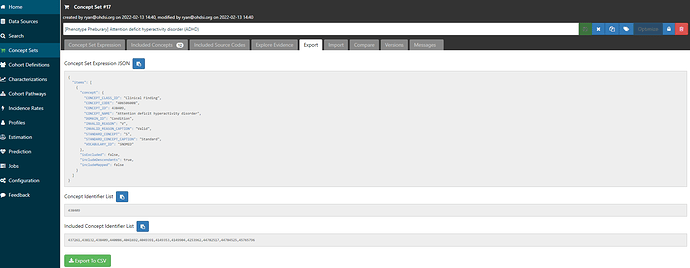
So, I’ll export this Included Concept list to go back to PHOEBE:
When I paste in this concept list into PHOEBE’s Concept Set Recommender, and ‘Show recommendations’, then I see the following set of concepts to consider:
We take some solace that the first 4 concepts shown are already included, so, at least on the surface, it doesn’t feel like we’re missing any big hitter concepts. #5 and #6 on the recommendation list are ‘Not included - parent’, which means these are concepts of something that is in the included list. ‘Disorders of attention and motor control’ seems - based on the label alone - like it could be too broad of a clinical idea, not specific enough for ADHD. Plus, PHOEBE tells me that there are only 7 databases that contain this concept, so I’m not worried (yet). ‘Disorder in remission’, clearly that’s non-specific, so we ignore that recommendation. #10 is ‘Not included - recommended via standard’ : ‘Adult attention deficit hyperactivity disorder’, well, that’s a concept that I surely want and I’m surprised that it doesn’t actually roll up to ADHD in the SNOMED hierarchy, but nevertheless, I’ll add that one to our conceptset expression. #14 is also ‘Not included - recommended via standard’, and its a procedure, ‘Drug therapy for attention deficit hyperactivity disorder’, that’s an interesting one insofaras if I’m looking to find a person with ADHD, then a person with an observation of taking a drug for ADHD probably has ADHD, so I’ll add this to my list. #15 is ‘Hyperkinetic syndrome with developmental delay’, and I’ll pull this in also since ‘Hyperkinetic conduct disorder’ is part of our current list. Feeling good about my new conceptset (so far).
Now, if I were to follow the guidance I’ve posted in prior threads, I’d now create a cohort definition using the conceptset in ATLAS, run jump out of ATLAS and into R to run CohortDiagnostics or PheValuator, and hope for the best.
But, lemme instead show a different trick that we can use, staying directly within ATLAS. As I mentioned in the clinical description, ADHD can be managed through pharmacologic treatment. And for the most part, ADHD drugs are most only indicated and prescribed for ADHD (with some use for nacrolepsy). (I reviewed the US FDA approved product labels for each of the drugs on DailyMed just to be sure) So, if we consider ADHD treatment as a potential marker for ADHD, then we can create a cohort of new users of ADHD drugs and characterize those patients to see what codes actually get used in the real world prior to treatment.
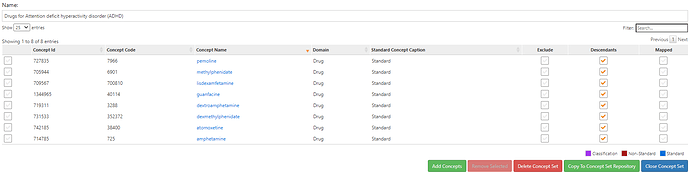
Here’s a conceptset of ADHD drugs:
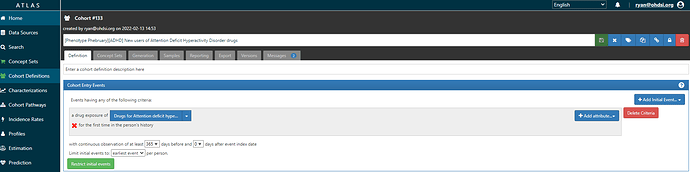
And a very simple cohort definition for new users of ADHD drugs (here’s the ATLAS-phenotype link):
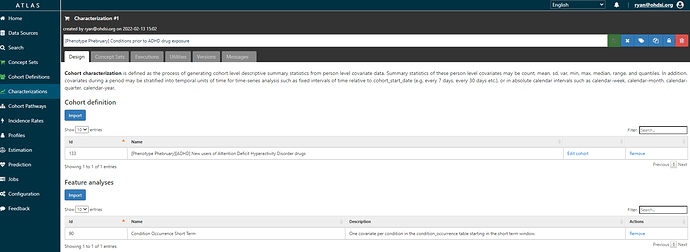
Then, with that cohort definition in hand, we can use the ‘Characterizations’ tab in ATLAS to produce a descriptive analysis about those qualifying patients. Here, all I want is to know what condition occurrence records are observed in the 30 days prior to or on index date, because I’m thinking those may be markers indicative of the indication. Here’s the Characterization analysis design (and link to ATLAS-phenotype as reference):
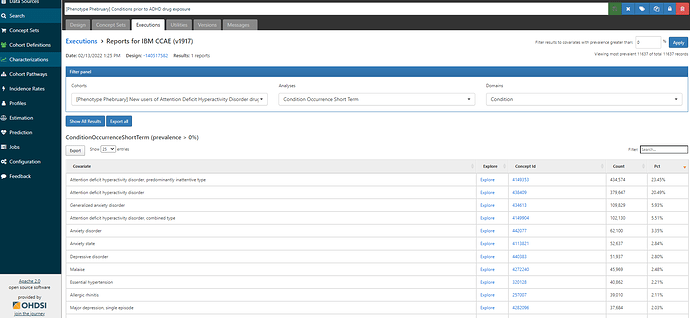
Here’s a table of results from the MarketScan CCAE database, a claims database from US:
As we see, the most prevalent of all condition concepts is ‘Attention deficit hyperactivity disorder, predominantly inattentive type’, occurring in 23.45% of the new user ADHD drug cohort. We can see the second concept is ‘Attention deficit hyperactivity disorder’, observed in 20.49% of the cohort, and the fourth concept is ‘Attention deficit hyperactivity disorder, combined type’ at 5.51%. All three of these concepts are in our existing conceptset expression for ADHD, so I’m feeling good (for now). I’ll note that we see ‘Generalized anxiety disorder’, ‘Anxiety disorder’, ‘Anxiety state’ as the other common concepts, which helps provide context for other comorbid conditions or differential diagnoses. But, in this case, I know I don’t want to include anxiety-related concepts in my definition so no action to be taken. Note also, we don’t see any other concepts occurring more than 3% of persons, so this gives us good perspective that we are unlikely to be missing a major condition concept that is hiding all the indicated patients.
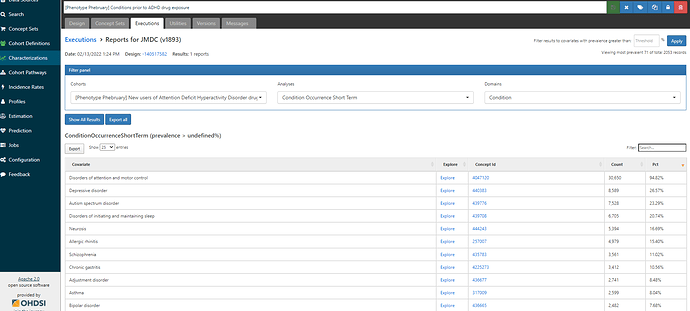
Now, let’s look at the results from JMDC, a claims database in Japan:
Totally different story here. We see there is one concept, ‘Disorders of attention and motor control’ that is observed in 94.82% of the ADHD drug new user cohort. There are NO concepts for ‘attention deficit hyperactivity disorder’ or any of the concepts we had in our draft conceptset. Remember, PHOEBE recommended ‘Disorders of attention and motor control’ earlier, but I dismissed it because the label sounded to broad and non-specific. Well, the label may be broad and non-specific, but in JMDC, that concept is the ONLY place where we can find ADHD patients. Failure to include that concept will result in having extremely low sensitivity. But here’s the upside: inclusion of that concept doesn’t impact the US database, because that concept isn’t observed.
Where does this leave us? By using PHOEBE recommendations and complementing that with what we learn from an ATLAS characterization of a treatment cohort, we can create a conceptset that can work for different databases. And even though the specific concepts that get used in each database may be different, there is no harm caused by creating the composite (and yet, we get the value of standardization).
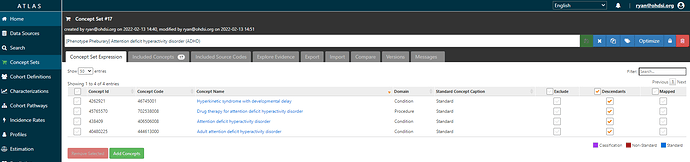
The final resulting ADHD conceptset?
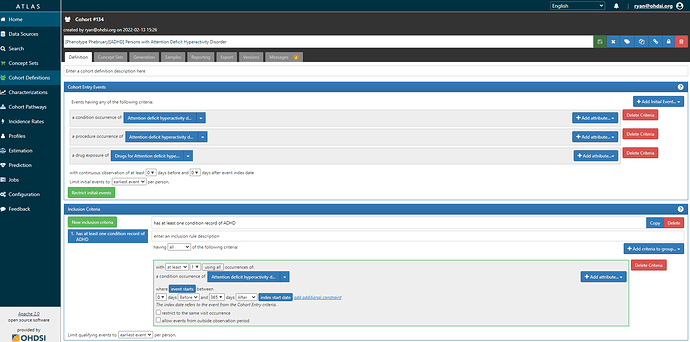
And here’s the cohort definition, note that we have to look for both conditions and procedures. Also note, we use the ‘index date re-calibration’ trick of allowing drug to be the first entry event so long as that is followed by ADHD diagnosis in the next year (ATLAS-phenotype link here):
My major ‘a ha’ from this experience: sometimes you have to let the data tell you what is happening in the real world, rather than you telling the data what you are looking for. In this case, had we not observed that patients in JMDC with ADHD meds didn’t have ADHD diagnoses (but instead, nearly all had a broader category), we could have had a definition that worked well in US but gave us grossly misleading results in Japan. In the context of OHDSI network studies, this is why we find it so critical to evaluate all cohort definitions within each data partner (and try to facilitate that process through CohortDiagnostics), because it can be the case that a definition that is deemed sufficient in one source may not generalize to another database. The more we have OHDSI network-wide resources, such as the Concept Prevalence results underlying PHOEBE, the more we can mitigate the risks of these errors occurring, but we still need to check in each source in each study to make sure our phenotype definitions are acceptable across all participants.