In this edition to Phenotype Phebruary, I’d like to discuss the work @Jill_Hardin and I did for developing phenotype algorithms in the immunology space for systemic lupus erythematosus.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease of unknown origin. Clinical manifestations include fatigue, arthropathy, and involvement of nearly all organ systems, particularly cardiac and renal.(Jump et al, Greco et al, Miner et al, Danila et al) A review by Stojan and Petri of research on multi-country incidence rate estimates found the incidence rate of SLE to be between 1-9 cases per 100,000 person-years (PY).
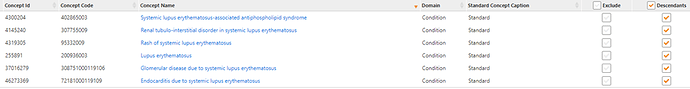
As @Patrick_Ryan has provided an excellent review of the details of the phenotype algorithm development process, I’ll build on that to demonstrate how we used the process for our cohort definitions. We first conducted a literature search for phenotype algorithms for SLE. From those resources we determined the codes used in prior studies. We used those as a starting point and entered those into the wonderful PHOEBE tool developed by @aostropolets . The final concept set was:
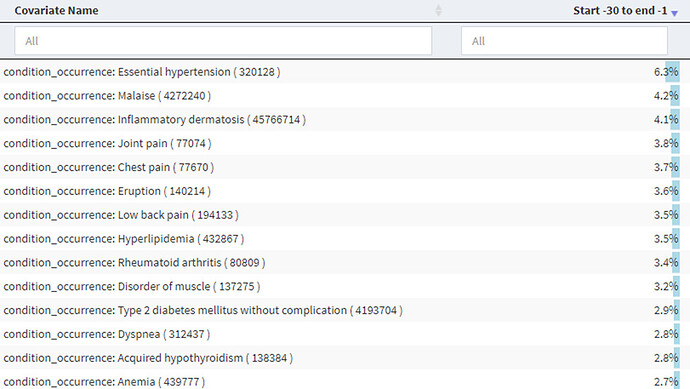
We then began building our cohort definitions. We were concerned about possible index date misclassification as prior research had indicated that there may be a long period between first symptoms and first diagnosis. We used the spectacular Cohort Diagnostics tool (thank you @Gowtham_Rao!) to examine the conditions and drugs in the time prior to an initial diagnosis of SLE and found in the IBM Commercial Claims and Encounters dataset:
We sorted through some of the big hitters, proportion-wise, across the different data sets and picked out ones that were likely candidates for SLE signs and symptoms and came up with:
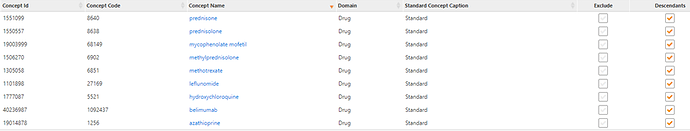
We did the same for drug exposures and developed:
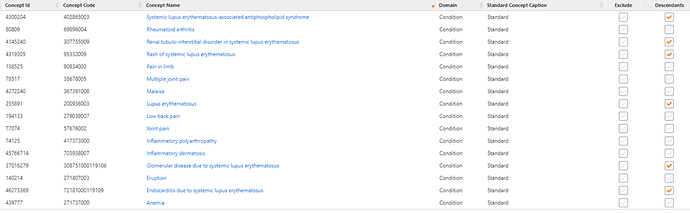
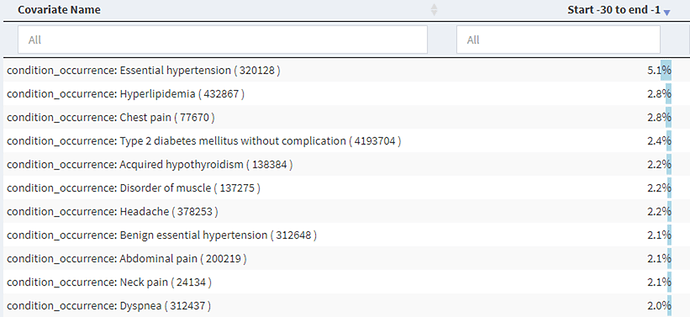
We developed cohort definitions for index date correction using 90 days as a possible time prior to the first diagnosis. In this definition, subjects would be included in the cohort if they had any of the signs, symptoms, or treatments as long as it was followed by a diagnosis code for SLE within 0-90 days after the sign, symptom, or treatment. Using this approach, the corrected index date is the date of occurrence of the sign, symptom, or treatment or an SLE code, whichever came first. Using this definition, we saw a reduction in the possible signs and symptoms in the 30 days prior to this new index date:
[NOTE: here is the link to the the Cohort Diagnostics shiny] We picked 90 days as a reasonable new index point though symptoms may occur for a longer time prior to actual diagnosis.
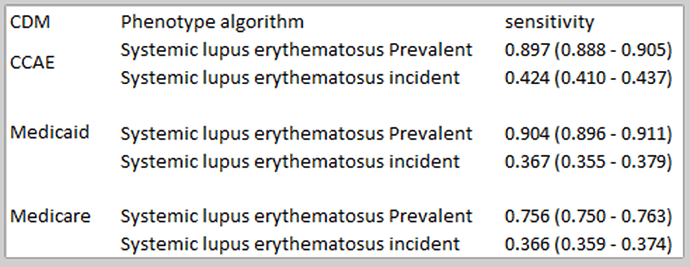
In our first set of definitions, we ran PheValuator and we noticed a big drop in sensitivity when we did not include a 365 day washout period prior to index in the analysis:
This begs the question of “what is the sensitivity estimate we want to use?”. In previous posts in the Phenotype Phebruary series, I’ve shown the results from PheValuator analyses where I did include the 365 day washout period and found high sensitivities. But I pose the question to the group if this is the way we want to look at sensitivity? To me, sensitivity measures how many subjects you found compared to how many you missed. If you impose a prior observation period of 365 days, you are surely missing subjects as the data above shows. If we impose a washout period in the analysis, that measure of sensitivity should contain the caveat that it is the sensitivity of the cohort definition of those with at least that length washout period prior to index. As they used to say on one of the TV talk shows – Discuss!
We next wanted to see if we needed to improve the specificity of the algorithm. For a single condition code cohort definition, we saw in Cohort Diagnostics:
That seemed like a large drop-off in the proportion of those with an SLE code in the time after index, so we developed a more specific definition requiring a second diagnosis code 31-365 days after index and found:
Which showed a nice increase in the in the proportion of those with an SLE code in the time after index. The measure we looked at was in the 1-30 day post index as the 31-365 day measure included required codes in cohort C2.
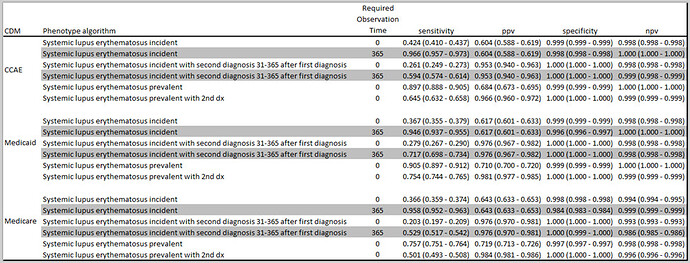
We ran PheValuator a second time and found:
We see the large drop in sensitivity when we compare the incident cohorts analyzed with 0 days prior washout to the ones with 365 day prior washout (gray shaded lines). We also see a large increase in positive predictive value (PPV) when we compared a single code algorithm to an algorithm requiring a second code. This, though, came at a cost in sensitivity.
In our final analysis, we concluded that we should make four algorithms available for use with different “fit for function” purposes. If your research requires a high sensitivity algorithm, possibly looking at SLE as a safety outcome where you don’t want to miss cases, then you might consider cohort definition “Systemic lupus erythematosus prevalent and correction for index date”. If you want to ensure that you have prevalent cases of SLE with a high probability of truly having the condition, for instance in drug comparison studies where you want only those subjects with SLE prior to drug exposure, you may want to use “Systemic lupus erythematosus prevalent with 2nd dx and correction for index date”. If your research requires incident cases of SLE, then you may want to use either “Systemic lupus erythematosus incident and correction for index date” if you want to maximize sensitivity or “Systemic lupus erythematosus incident with 2nd dx and correction for index date” if maximizing specificity is your requirement.

 )
)