Great conversation.
1. Regarding re-indexing: @Patrick_Ryan regarding re-indexing - in the video recording (see link) we discussed this. @allanwu made a point that the operating characteristic he is looking for in current iteration was specificity and PPV. Index date misspecification error was acceptable at this time. Also the intent was to replicate a published algorithm, which was indexing on latest event. In the group discussion, it was decided to explore reindexing after achieving these above goals.
2. Regarding reindexed cohort definition developed by @Patrick_Ryan : Btw @allanwu i fixed this
3. Regarding comparing Tiered definition with unaminity algorithm:
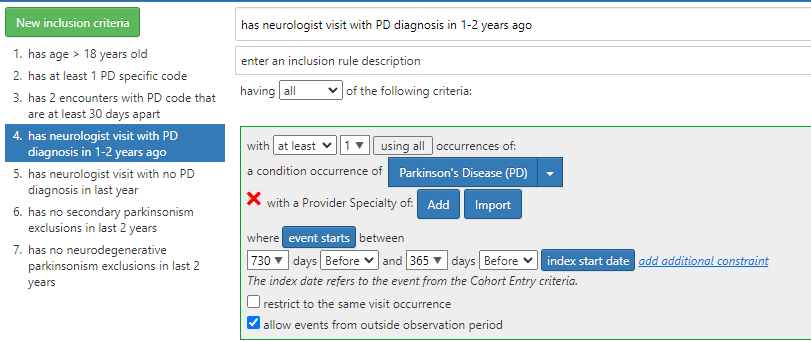
This is purely technical - we cannot execute a cohort definition when the cohort definition is incomplete i.e. provider specialty. i.e. i cannot re build cohort definitions without this issue being fixed
4. Regarding bugs in software at data.ohdsi.org/PhenotypeLibrary . The issue appears to the server infrastructure running the OHDSI tool. The error in incidence rate plot (tagging @lee_evans and @jpegilbert has been previously reported but I think there were some challenges in fixing it