@overevkina
There is an OMOP CDM Oncology Extension that is currently being worked on. We are in the process of completing some vocabulary and ETL documentation work to be able to test the extension. We are meeting weekly. Meeting info here:
https://www.ohdsi.org/web/wiki/doku.php?id=projects:workgroups:oncology-sg
Pleas join us.
Here is an overall explanation/discussion of the extension:
https://github.com/OHDSI/CommonDataModel/files/2642492/Oncology.CDM.Proposal.2018-12-02.pdf
https://github.com/OHDSI/CommonDataModel/issues/239
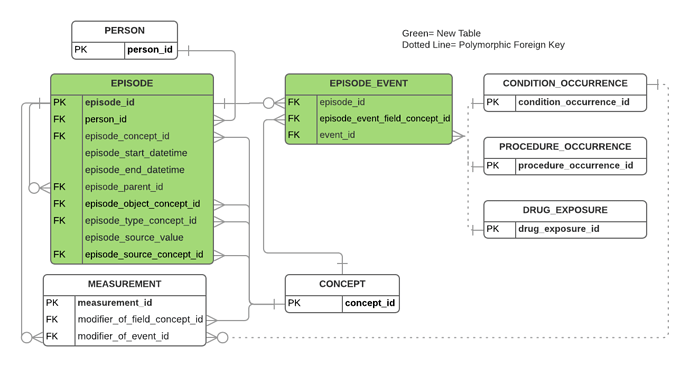
Here is the extension data model:
The main idea of the extension is to add an EPISODE table that can represent a base oncology diagnosis and to add modifier_of_event_id and modifier_of_field_concept_id fields to the MEASUREMENT table to allow for the refinement of an oncology diagnosis with modifying tumor characteristics (MEASUREMENT becomes a polymorphic child of EPISODE).
We want create a place to track the progression of an oncology diagnosis over and above the problem list/claims observational welter of the CONDITION_OCCURRENCE table. This welter can be linked to via the EPISODE_EVENT table.
We have added the International Classification of Diseases for Oncology (ICDO) standard for the base representation of an oncology diagnosis: site/histology or topography/morphology. See here: https://codes.iarc.fr/.
An ICDO site/histology pairing (pre-coordinated if possible to SNOMED) should be placed in the EPISODE.episode_object_concept_id. The EPISODE.episode_concept_id field should contain the state of the ‘Disease Episode’. For example, ‘Disease First Occurrence’, ‘Disease Recurrence’, ‘Disease Progression’. Linking between ‘Disease Episode’ entries can be accomplished via parent/child relationships between Episodes via the EPISODE.episode_parent_id column.
We have added the NAACCR tumor registry data dictionary as the initial standard vocabulary of modifying tumor characteristics, e.g., staging, grade, lymphatic invasion, tumor size. See here: http://datadictionary.naaccr.org/default.aspx?c=10. We are looking at the possibility of also adding the CAP cancer checklists as another vocabulary of modifying tumor characteristics. See here: Cancer Protocol Templates | College of American Pathologists
Looking at your example. We have been concentrated on pathology-focused tracking of tumors. I think you are doing imaging-focused tracking of tumors. I don’t think NAACCR or CAP covers some of the attributes you list. So you could help us enrich our extension by better supporting imaging-based tumor findings.
Taking your example with my annotation in bold:
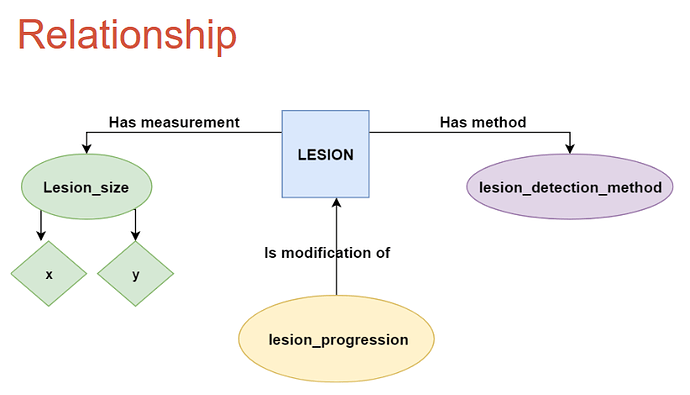
- lesion_site (brain) EPISODE.episode_object_concept_id you. You are “missing” histology/morphology. Do you have that? For example anaplastic astroctytoma or glioblastoma? Maybe not if this is pre-surgical monitoring.
- lesion_location (cerebellar) EPISODE.episode_object_concept_id
- lesion_orientation (left) MEASUREMENT pointing to EPISODE
- lesion_detection_method (x-ray) MEASUREMENT pointing to EPISODE
- lesion_size MEASUREMENT pointing to EPISODE
- lesion_size_x MEASUREMENT pointing to EPISODE
- lesion_size_y MEASUREMENT pointing to EPISODE
- lesion_recist (bool) ?
- lesion_recist_responce (stable diseases) EPISODE.episode_concept_id with a concept of ‘Stable Diseases’ and EPISODE.episode_parent_id pointing to prior state of the “lesion”
- lesion_recist_size MEASUREMENT pointing to EPISODE
- lesion_recist_size_x MEASUREMENT pointing to EPISODE
- lesion_recist_size_y MEASUREMENT pointing to EPISODE
- lesion_recist_detection_method MEASUREMENT pointing to EPISODE