Disclaimer: This post advocates for increased use of the FACT_RELATIONSHIP table and may not be suitable for all viewers.
Problem: There is a growing list of disparate use cases that require persisting relationships between clinical events to which the existing foreign keys are insufficient.
I view the overall goal simply as establishing a common convention. Not everyone will have this relational information explicitly in their data but for those that have use cases and wish to preserve it in OMOP, merely settling on a standard approach as to how to store it to enable interoperability is sufficient.
Some of the use cases:
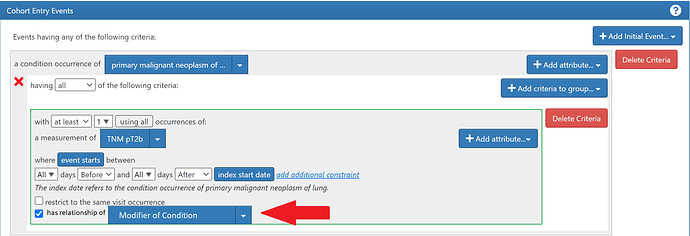
- details (modifiers) of conditions (staging, topography, subtype, findings, progression, etc.)
- details (modifiers) of procedures (margins, complications, etc.)
- linkage between NLP derivation and resulting clinical event records
- providing context as to what procedure/test/device exposure lead to an observation or measurement (most notably from derived data) e.g., interpretation finding from a specific type of imaging; a genomic measurement from a specific type of panel, etc.
- a condition resulting from another condition (e.g., metastasis)
- a procedure targeting specific condition (e.g., therapy targeted to a specific lesion)
- A drug or procedure as part of a regimen/treatment plan
- Temporal, one:many relationships when the model constraints require 1:1, such as patient or care site locations over time, patient to provider relationships over time, etc.
This overall need has been incrementally enabled on a use case based approach, growing organically.
Current Approach: Two-part foreign key
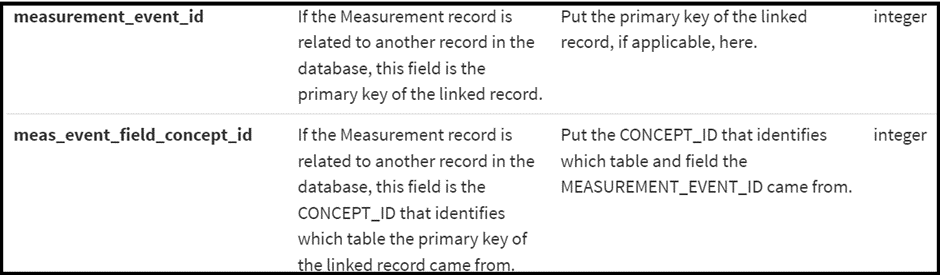
To enable storing attributes (modifiers) of conditions and procedures, two fields were added to the MEASUREMENT table. This was initially implemented with the complexities of oncology data in mind.
This mechanism can be thought of as a two part foreign key { table identifier, row identifier } which allows the flexibility to attribute a MEASUREMENT, or modifier, to any row of any table.
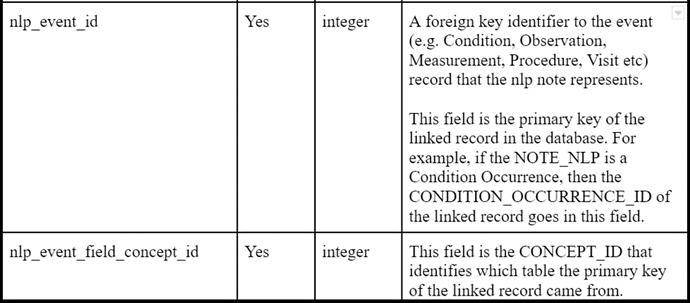
Additionally, the EPISODE_EVENT uses this same mechanism to link an EPISODE to any other clinical event.
And, as was recently ratified in the CDM WG, the same mechanism will be added to NOTE_NLP to provide provenance to NLP derived events in other tables.
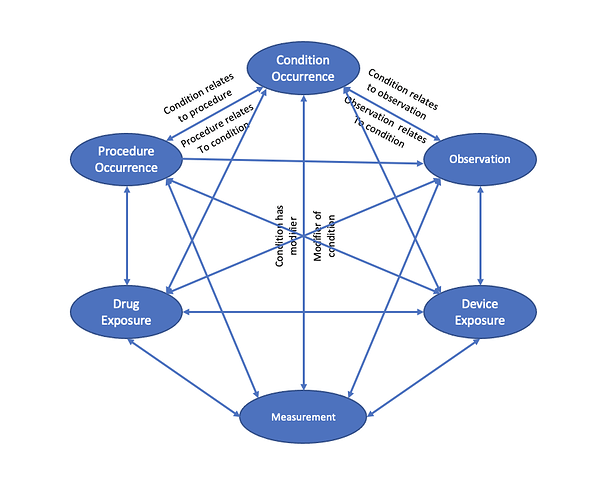
This approach is useful but there are three notable deficiencies:
- In terms of extensibility, to fully encapsulate the spectrum of relationships between clinical events from existing and emerging use cases, we would need to add these same two columns, redundantly, to nearly every table. I’m sure we could come up with an example for every table to table combination
- There is no context as to what type of relationship exists. It is simply a foreign key; “this is related to that”
- There is the potential to run into a recursive situations, making it difficult to functionally account for. E.g. “a measurement of a measurement of a measurement” – at what point do the functions stop checking for additional relationships?
After running into the issue of wanting to create a relationship from one condition to another condition (i.e. representing a metastasis with it’s own distinct attributes), I dug into the possibility of a more extensible and reliable approach, described below.
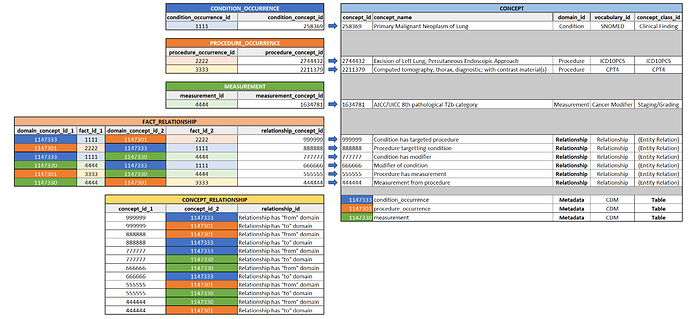
Approach 2: FACT_RELATIONSHIP table
Let’s look at the fact relationship table:
- Currently, it seems to mostly be used for relationships from one person to another, but has flexibility for much more
- Essentially, it has the ability to cover all of the above use cases without needing to append addition columns to the event tables.
- It provides the missing “context” as to what the relationship is
- From a downstream perspective, I would suppose this approach would be easier to integrate with existing OHDSI tooling vs. needing to check every table’s FKs
- The presence of a concept to represent the type of relationship allows for mechanisms of validation and tooling
The “relationship” domain represents what can be stored in this “relationship_concept_id”. An example is :“aunt of subject”
If we were to leverage the FACT_RELATIONSHIP structure and add a set of robust concepts with functional relationships to contextual concepts, it could be an extensible approach applicable to the entire spectrum of use cases mentioned above.
For example, using a couple of the use cases above, adding both a “to” and “from” domain relationship for validation/DQ check purposes:
| Domain ID | Concept Class ID | Concept Name | (Relation)Has concept_id_1 Domain | (Relation)Has concept_id_2 Domain |
|---|---|---|---|---|
| Relationship | Attribute | Attribute of Condition | MEASUREMENT | CONDITION_OCCURRENCE |
| Relationship | Attribute | Condition has attribute | CONDITION_OCCURRENCE | MEASUREMENT |
| Relationship | NLP derived | Observation from NLP | OBSERVATION | NOTE_NLP |
| Relationship | NLP derived | Note has observation | NOTE_NLP | OBSERVATION |
Temporality
To take things a step further, there are a handful of other use cases that have been discussed, requiring creation of new tables, that could be covered by the same structure if temporality is included. Notably, the representation of data where the model forces a 1:1 relationship and the source data is often one:many, such as:
- Person to location; Care_site to location
- Person/provider to care_site
- Person to provider
If we wanted to kill a dozen or so birds with one stone, we could then add start and end date to FACT_RELATIONSHIP, as optional fields, which would additionally cover these use cases.
Something along the lines of:
(Revised) FACT_RELATIONSHIP
| CDM Field | Datatype | Required |
|---|---|---|
| domain_concept_id_1 | integer | Yes |
| fact_id_1 | integer | Yes |
| domain_concept_id_2 | integer | Yes |
| fact_id_2 | integer | Yes |
| relationship_concept_id | integer | Yes |
| start_date | Date | No |
| end_date | Date | No |
And perhaps for further validation/DQ purposes, the relationship concept could have a relationship to a context concept to indicate whether it is temporal in nature or not.
E.g.
| Domain ID | Concept Class ID | Concept Name | Has concept_id_1 Domain | Has concept_id_2 Domain | Has relationship type |
|---|---|---|---|---|---|
| Relationship | Location History | Resided at location | PERSON | LOCATION | temporal |
| Relationship | Location History | Had resident | LOCATION | PERSON | temporal |
| Relationship | Provider History | Had PCP | PERSON | PROVIDER | temporal |
| Relationship | Provider History | PCP of patient | PROVIDER | PERSON | temporal |
The above example is one approach but the “is temporal” context could be handled in other ways, such as determined by the domain or concept class.
Implications:
- The altered approach seemingly simplifies and futures proofs the CDM for the existing and emerging use case demands
- Can revert back to standard CDM and remove the additional columns from MEASUREMENT and NOTE_NLP
- Eliminates the need for the tables: EPISODE_EVENT, LOCATION_HISTORY (as well as proposed others such as PROVIDER_HISTORY, CARE_SITE_HISTORY, etc.)
- Adds ability to distinguish between different types of relationships within the same domain pairs (e.g. “patient lived at location” vs. “patient works at location”)
- Not to open another large can of worms at the same time but, there have been developing discussions of a need to record the “derivation method” for certain types of data where the type concept alone is not sufficient. For example, the specific NLP, drug regimen derivation, episode derivation, geospatial algorithms used to derive the data. This is helpful to adequately preserve the provenance when comparing data across sites in studies. Should that become a reality, that we have something along the lines of a “derivation” table, it could seemingly easily slide into this same mechanism
Unknowns:
- Regarding integration with existing tooling, what are the implications between the two approaches. As far as I know, none of the “two part foreign key” functionality has been built into any ETL tooling (e.g. Perseus), ATLAS/WebAPI, or HADES. Neither approach is entirely straightforward for ETLs given the complexity of the nature of inserting into identity tables and preserving the relationship using those keys, but I’d be curious to hear from the developers of the above if one or the other approach is viewed as more feasible. @schuemie @anthonysena @bradanton
- Efficiency wise, I would imagine that moving all of these relationships into a single table could be an efficiency concern, but it isn’t clear if that is valid or not given the FACT_RELATIONSHIP table would consist of indexed integers
- Whether or not I will be burned at the stake for this FACT_RELATIONSHIP heresy
Thoughts?