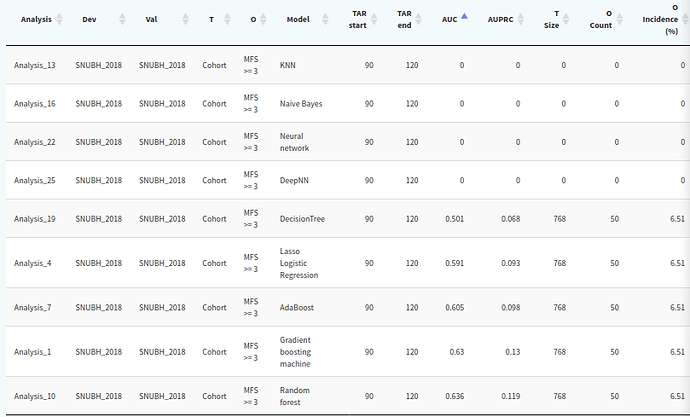
I tested patient level prediction package.
Empty cohort issue only occurred on KNN, naive bayes, neural network, deep NN model.
Decision tree, lasso logistic regression, ada boost, gradient boosting machine, random forest model work normally.
All models used default option.
The plplog of KNN model is as below.
|2019-12-17 10:53:45|[Main thread]|INFO|PatientLevelPrediction||Patient-Level Prediction Package version 3.0.8|
|2019-12-17 10:53:45|[Main thread]|INFO|PatientLevelPrediction|runPlpAnalyses|No plpData - probably empty cohort issue|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Patient-Level Prediction Package version 3.0.8|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||AnalysisID: Analysis_13|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||CohortID: 19|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||OutcomeID: 501|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Cohort size: 3080|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Covariates: 1621|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Population size: 3080|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Cases: 204|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||testSplit: subject|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||outcomeCount: 204|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||plpData class: plpData|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||testfraction: 0.25|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||nfold class: numeric|
|2019-12-17 13:05:06|[Main thread]|DEBUG|PatientLevelPrediction||nfold: 10|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||splitSeed: 1935504|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction|subjectSplitter|Creating a 25% test and 75% train (into 10 folds) stratified split by subject|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction|subjectSplitter|Data split into 768 test cases and 2312 train cases (232, 232, 232, 232, 232, 232, 232, 230, 230, 228)|
|2019-12-17 13:05:06|[Main thread]|INFO|PatientLevelPrediction||Training KNN model|
And used covariates are as below.
Demographics Gender
Demographics Prior Observation Time
Demographics Post Observation Time
Demographics Time In Cohort
Drug Era Long Term
Procedure Occurrence Long Term
Measurement Long Term
Measurement Value Long Term
Observation Long Term
Charlson Index
Dcsi
Chads2Vasc
Hfrs
Distinct Condition Count Long Term
Distinct Ingredient Count Long Term
Distinct Procedure Count Long Term
Distinct Measurement Count Long Term
Distinct Observation Count Long Term
Visit Count Long Term
Visit Concept Count Long Term
How to solve this problem?
Thank you.