Glad that it worked for you 
Yes, those will be the steps. But I haven’t yet figured out connecting/updating/extracting to and from ‘in memory’ database yet.
Folks - great work so far. Seems @alexander and @rohitv have been busy.
We discussed on last week’s call that we do not want to reinvent the wheel. There was strong agreement that we want to use the OHDSI Methods Library where possible.
I propose we have a call this week to talk through the task list (based on the prep work people have been testing out this week) and use that time to outline what needs to get done in what order. From there we can start having people sign-up and own different parts, so work can be distributed.
I sent an invite to those in the Google Doc. Here is the meeting link for those who want to join:
https://iqvia.webex.com/iqvia/j.php?MTID=m4168ad2e0610df768dec9900f4fabef5
The call will be held tomorrow, October 3 from 12-1PM EDT.
Hi @krfeeney . May I ask you to shift it on one hour earlier or later, please? I’m busy 9:00 - 10:am PST on weekdays and can’t move it. Thanks.
Do we want to do the hour earlier? It is extremely late for South Korea.
8:00 am PST works for me.
Regards
- Rohit
@krfeeney I’m okay, I will rest well in the morning because today is holiday in Korea. 
I am also studying Docker now. I’m trying to run the docker image, but since it’s a windows environment, it doesn’t work. I’ll find a solution today.
Another thing to note is that if you use synapse as a Google account, you need to register an account password in advance. I’ll summarize it in the github issue.
Dear all the Erasmus MC Health Data Science group would like to join this team.
@Rijnbeek, @RossW, @lhjohn, and my new student Cynthia Yang.
Will will join the team also on the website. @krfeeney
Welcome @Rijnbeek, @RossW, @lhjohn and Cynthia! We’re delighted to have your expertise on board.
We’re trying to have a quick touch point to help organize some of the chatter that’s been on this post this week. We’ve moved the teleconference up to 8AM PST / 11 AM EST TODAY (3 Oct 19). The WebEx link is: https://iqvia.webex.com/iqvia/j.php?MTID=m4168ad2e0610df768dec9900f4fabef5
Audio Information:
US Call-In: 1-844-517-1271
Korea Call-In: 00798-1-1-003-7282
Netherlands Call-in: 08000201661
Universal Passcode: 964100974
@rohitv @alexander - I’ve got a call during the first 30 mins of this time but don’t wait for me because it seems like it works for everyone else. Can I ask you two to start off the conversation and capture a few notes in the Google Doc? I’ve put an agenda in our Google Doc: https://docs.google.com/document/d/1y0lkFRKOiDrV6D9Cm5PYx-_NSVwWfIvekKTE_xprQGs/edit?usp=sharing
Once I join, I can take the action to consolidate the output and get the task list divided up with timelines.
What did you plan to do? Sorry, I had to jump to another meeting at 10 am. I can help with populating CSV files into SQL database, but I have some time to do it only next weekend. I believe making a PLP framework working with the data format they provide us is on a critical path now and blocking almost everything else. Is it correct?
I was also thinking that DatabaseConnector also works with SQLite. And puting the CSVs into SQLite may be easier. Puting in postgres is not a problem but HAVING the postrgres server itself running fine on the Syapse platform is I think the key problem.
I found this code snippet: https://stackoverflow.com/a/28802613/984532
This can be a good option too http://hsqldb.org .
But I would prefer to stay with Postgeres as long as possible to avoid issues with PLP framework. I don’t think database will be a bottleneck for what we are doing.
I’m very skeptical that the resources they give per submission are enough to train a good model.
We should try it, but I believe finally we’ll come up to using their data only as dev and test datasets according to Andrew Ng definitions https://cs230-stanford.github.io/train-dev-test-split.html (According to Andrew’s courses it’s OK to have different data distribution between Train and Dev datasets. But the distribution of data in Dev and Test datasets must be the same)
Even if we figure out how to do training for 1 hour on a 4core CPU machine without GPU they have another constraint: 5 submissions per day… It’s not enough to do hyper-parameter optimization for the ML model…
All - we came up with this list of tasks… now we need people to pick up action items.
| Task | Owner | Can this be done concurrently? | Status |
|---|---|---|---|
| Docker image can install PLP package | Rohit / Alexander | n/a | Complete |
| Docker image can connect to DB | Rohit / Alexander | n/a | Complete |
| Write R script to read CDM files | To be assigned | Yes | Not started |
| Populate CDM on database (e.g. Postgres) | To be assigned | Depends on prior step | Not started |
| Write Target cohort query (e.g. in JSONs) | To be assigned | Yes | Not started |
| Write Outcome cohort query (e.g. in JSONs) | To be assigned | Yes | Not started |
| Ingest cohort JSON using PLP framework | To be assigned | Depends on prior step | Not started |
| Run feature extraction from cohort JSON | To be assigned | Depends on prior step | Not started |
| Run a model from cohort JSON | To be assigned | Depends on prior step | Not started |
| Round 1 Submission: Complete full cohort run (note: don’t focus on run time or AUC) | To be assigned | Depends on prior step | Not started |
If you want to pick up a task, you can reply and then put it in the google doc: https://docs.google.com/document/d/1y0lkFRKOiDrV6D9Cm5PYx-_NSVwWfIvekKTE_xprQGs/edit
Once we know who owns what, we’ll set-up periodic check ins over the next 4 weeks. There is active discussion on the resourcing constraints that requires some testing of the PLP framework to understand what’s possible. Note, we agreed this is not the focus of Round 1.
Thank you for always @krfeeney 
I’ll take these :
- Ingest cohort JSON using PLP framework
- Run feature extraction from cohort JSON
@alexander and @Chungsoo_Kim added these to the Google Doc. 
I believe @jswerdel is working on a quick test to see the run time of PLP in his environment. Going to put that in the grid too so it’s captured.
Do we want to do another check in towards the end of next week?
I am okay with a call by the end of next week to touch base on the progress.
Regards
-Rohit
Hello Everyone,
Can I seek your help on the below issue?
I am unable to push the docker image by following their tutorial (Synapse | Sage Bionetworks). I encounter the usual "denied: Requested access to the resource denied"
I see that this issue is very common and everyone had a trial and error approach to make this work.
However, I couldn’t get this working. Can you help me with this?
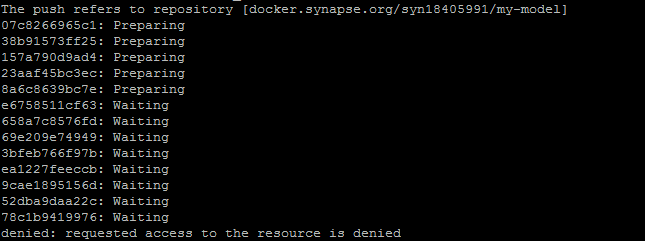
The command that I used are given below
docker login docker.synapse.org #login is successful
This is how my repository looks like
Your help is much appreciated
Thanks
Selva
Hi Selva. Are you member of our team on Synapse? It seems like you don’t have an access to the project. https://www.synapse.org/#!Team:3397227