Hello,
when running a study against Eunomia for testing purposes (using this specification, which doesn’t make sense clinically but was only created to retrieve counts in Eunomia), I am running into the error: ‘error calling function ‘fun’, no rows to aggregate’.
Is this something common? Maybe someone can help with where to look for the solution? The module itself still produces the cm_results file, but fails to produce the results folder.
Thanks!
Hi @TatianaSkugarevskaya ! Could you provide a bit more detail? At what point does the error generate? Do you have a log file and/or an error report? Code that was just to run the study?
Hi @schuemie , thanks for reaching out!
I am running the basic StrategusCodeToRun script for executing Strategus with the specs from my previous post, and the database I am connecting to is Eunomia. Again, this is a test run for the specs, and they only partially represent what I intend to use for the study itself (packages and settings are the same, cohorts are different, simpler). It is already apparent, though, that the error replicates with the ‘normal’ specs and non-synthetic databases too.
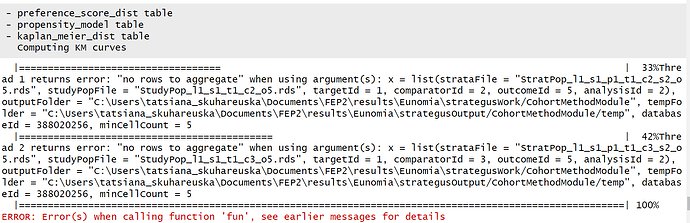
The Strategus package includes CohortGenerator, CohortDiagnostics and CohortMethod modules, and the fist two run correctly. In the CohortMethod part, however, the log shows only partial execution, with breaks after the ‘Computing KM curves’ portion, as on the screenshot.
As I understand, this might be the case if the strata are small (even though the cohort definitions were designed to have at least several hundred patients in Eunomia). My main question is: what is the reason for the error? And is this a systemic study specs error or can we expect it to fix itself with bigger patient counts?
Thank you!
I am sorry, but I am unable to reproduce that error. I have cloned ‘FepPharmacotherapy’, restored the included renv.lock file, and have executed StrategusCodeToRun.R on Eunomia, all without error.
It is already apparent, though, that the error replicates with the ‘normal’ specs and non-synthetic databases too.
Could you comment on this? We have many tests in place to make sure the code runs, also against Eunomia. It would be good to understand why you see consistent errors.
Which version of R are you using? What operating system? Was an error report generated, and if so, could you share it?
Thank you for looking into that! (and sorry for the delayed answer)
Did you use the specifications file used in the StrategusCodeToRun.R or the one from my initial post? The difference here is, the script runs without errors for me too if using the ‘full’ study specs, the ones mentioned in the ‘CodeToRun’ file. However, the cohort definitions used there are complex, and do not produce any patients in Eunomia. So I ‘made up’ the simpler, ‘test’ versions of cohorts in order to test the script with non-0 patient counts. And there the problem starts.
Here I meant that the error occurs whenever the script is being run with non-empty cohorts (either with the ‘test’ specs in Eunomia or with ‘normal’ specs in any other CDM). Basically, for some reason the error happens when there are patients within the cohort, and if there are none (as when running the ‘normal’ specs against Eunomia, due to the definition complexity), then everything runs smoothly.
I am using R 4.4.2 on Windows 10 64-bit. The log with errors from one of such runs is here.
Thank you. I missed the fact that there were two different specification files. Switching to the simpler one allowed me to reproduce the error.
The error is caused by the comparator cohort being empty while the target is not, in combination with PS stratification. This was not one of the scenarios in our unit tests, which is why I hadn’t noticed it before.
I have fixed the code so it doesn’t throw an error in this case, and updated our unit tests. If you already want to use this new version in your code you can run:
remotes::install_github("ohdsi/CohortMethod", ref = "develop")
I’ll probably create a patch release with this fix in the next few weeks.
Thank you, that sounds really promising!
I have tried running ‘test’ study with the dev package version, the error did not pop up, however, now it returns the following error
FATAL tidyselect Can’t extract columns that don’t exist. ![]() Column
Column targetAtRisk doesn’t exist.
Almost in the same execution section, when doing KM curves. The new error log is here.
Is this something you also expect to be fixed with the new release?
I’m sorry, I’m not able to reproduce that error using the new version of CohortMethod and inst/FepPharmacotherapyAnalysisSpecificationTest.json.
In the log file I see the tables already existed. Could you try running from scratch, including deleting the results folder?
Oh, for some reason it ran correctly today, maybe I missed something yesterday.
Anyways, thank you so much for your help, will be looking forward to the patched script version!