The CohortDiagnostics package (at GitHub - OHDSI/CohortDiagnostics: An R package for performing various cohort diagnostics. ) is very useful.
There are instructions, how to use it at a site that also has Atlas/webAPI installed.
Running cohort diagnostics using WebAPI • CohortDiagnostics
Those who use it in R, it would be good to have a vignette (or other guide) how to use it at a site without Atlas/webAPI installed.
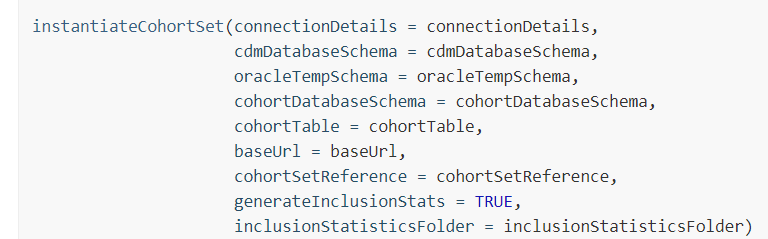
This code here https://github.com/ABMI/Covid19Characterization/blob/master/Covid19TestCharacterization/extras/CodeToRun.R#L46 seems to demostrate that a site does not need Atlas.
It seem like I just need to emulate this call using R code (and not webapi)
Is that right? The assumption is that I have .SQL definitions of the cohorts in my local hard drive. (like 2018 and 2019 network studies tended to do)
E.g. SkeletonDescriptiveStudy/CreateCohorts.R at master · vojtechhuser/SkeletonDescriptiveStudy · GitHub
Requiring a site to have webAPI fully working is a significant burden (block for some sites).