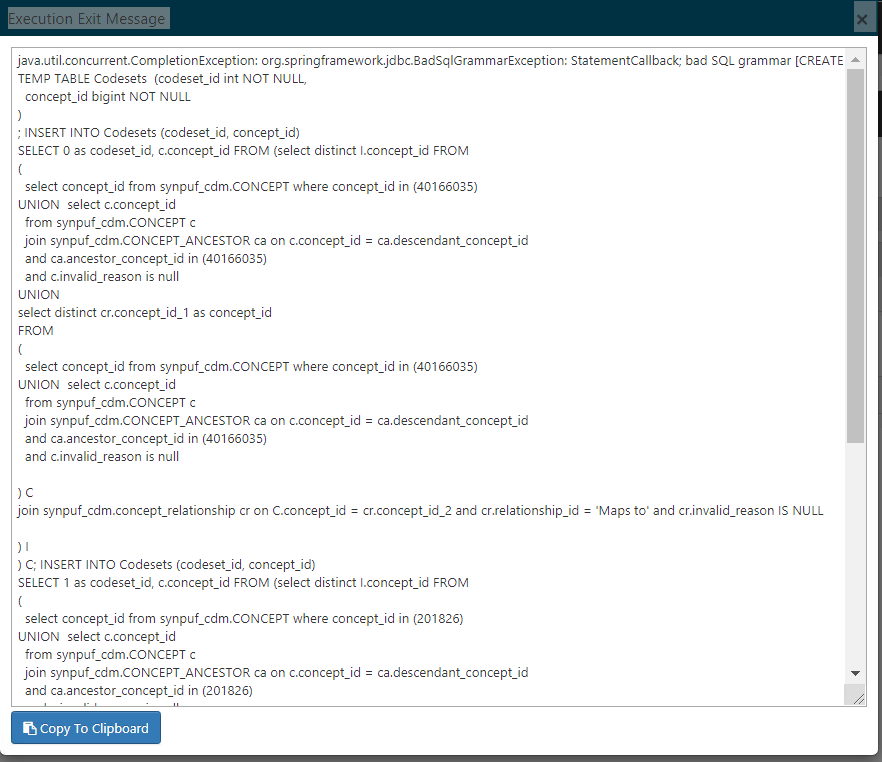
Hi All,
I am also facing a similar issue :
I am currently analyzing the features offered by the ATLAS tool. We are using a sample SynPuf data for this activity. We are using PostgreSQL based AWS RDS as the database.
Observation : The ATLAS tool created the cohorts and the reports were generated successfully. However, when we went to perform characterization and build cohort pathways, the process failed with the below error:
java.util.concurrent.CompletionException: org.springframework.jdbc.BadSqlGrammarException: StatementCallback; bad SQL grammar
Please refer to the image below :
We picked up the segment of queries mentioned in the error and executed it manually on our database and they executed without any issues.
We are looking for some help to resolve this. Please do let us know if you have any suggestions.
Thanks!