dear all,
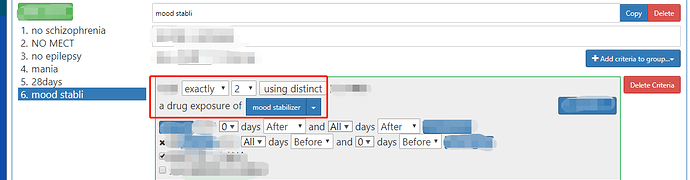
when I construct the cohort like the picture, exactly but when I search all the person using SQL in the database, I found there are still the person who only has one kind of drug in the cohort. why ? what is wrong?
Hi @pandamiao,
Quite hard to figure out the issue using just one screenshot.
Could you please share the concept set and your SQL ?
If you do narrow this down to a test case, I will add it to our suite of test-cases for Circe to ensure that this is working.
For now, I’ll just fabricate some test data in the test suite to ensure that the ‘exactly’ and ‘distinct’ options are working as expected.
thank you very much @Chris_Knoll. I search the person in the cohort in our own database, then find there is still the person who only has one kind of drug. do you have any idea about your test? thank you very much
thank you very much @Eldar, the sql
–
create table results.doubledrg as (
with ci as(
select ab.*,vo.visit_occurrence_id,visit_start_date from (
select * from results.cohort co where cohort_definition_id=283) ab
inner join beijing_omop_531_prd.visit_occurrence vo on ab.cohort_start_date=vo.visit_start_date
and visit_concept_id=9201 and visit_end_date-visit_start_date>=7 and ab.subject_id=vo.person_id
)
, de as(
select ancestor_concept_id, descendant_concept_id from beijing_omop_531_prd.concept_ancestor where ancestor_concept_id in ( 745466,751246,40058139,740275 )
)
,dru as (–7016条,742人
select ci.,de.,drug_concept_id,drug_exposure_start_date,drug_exposure_end_date,
dense_rank() over(partition by person_id order by ancestor_concept_id) as rank1 from ci ,de,beijing_omop_531_prd.drug_exposure deo
where de.descendant_concept_id=deo.drug_concept_id
and ci.visit_occurrence_id=deo.visit_occurrence_id
)
– select distinct visit_occurrence_id from dru
,sec as(–6515 ,658
select distinct * from dru where dru.visit_occurrence_id in (
select distinct dru1.visit_occurrence_id from dru dru1 where
rank1=2 ) order by subject_id,drug_exposure_start_date)
);
select distinct visit_occurrence_id from sec
In the UI, it gives you the complete SQL that you can execute manually and inspect the temporary tables that shows you the people that were identified as cohort entry, and then each inclusion rule (I’m assuming you’re using an inclusion rule for the ‘having exactly 2 distinct…’.
I’d suggest you run through he code manually and determine how it’s finding that person. To make it faster, you can even alter the code to limit to the specific person_id so that you don’t have to filter through lots of records to find the specific person.
My only guess is that you may have put a parent concept into your concept set, but it resolves into 2 children, and it is the children that are being coded in the record which is giving you the ‘multiple records from the single drug’. Even a single drug may have multiple drug forms (or dose strengths) and that would lead to 2 distinct counts.
you are right, @Chris_Knoll, I choose the concept id which is the ingredient and has many children. because I want to choose the subjects who eat only one kind of drug, the same ingredient.
In this situation, if the distinct cannot do this, how can I choose this person with atlas?
thank you very much
@pandamiao,
As far as I understood you want to include persons taking exactly 2 from different ingredients
( lamotrigine, Valproate, Lithium Carbonate, Carbamazepine)
If this is correct, than as a quick answer - you’d use Drug Era, not Drug Exposure.
Also, please make sure the concept set is defined on the ingredient level.
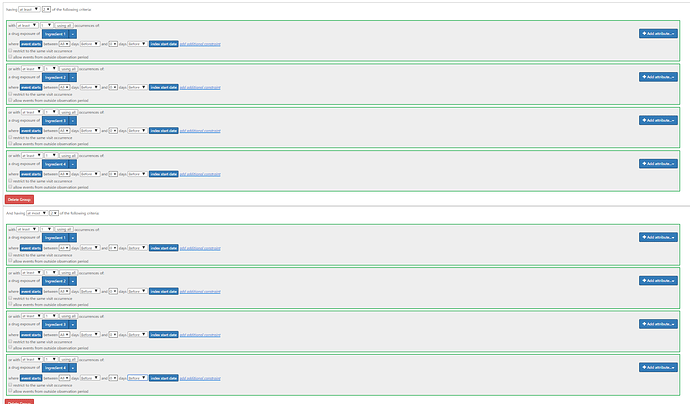
Another way of doing this to create separate concept sets for the four different ingredients, and use a ‘group criteria’ with the option to look for ‘at least X’:
You’d write it as:
Having all of
having at least 2 of (A OR B OR C OR D)
and having at most 2 of (A OR B OR C OR D)
(the at least 2…at most 2… means exactly 2…I don’t know why I didn’t put an ‘exactly 2’ in a group criteria, but it’s on my list to add it to the UI).
This is what it looks like in the UI:
Once the ‘exactly X in group’ is implemented, you wont’ have to duplate the ‘at least 2’ and the ‘at most 2’ statements.
But I also like @Eldar approach as well, that’s a good way to leverage the drug era table.
Thank you very much @Chris_Knoll, I will try this way.
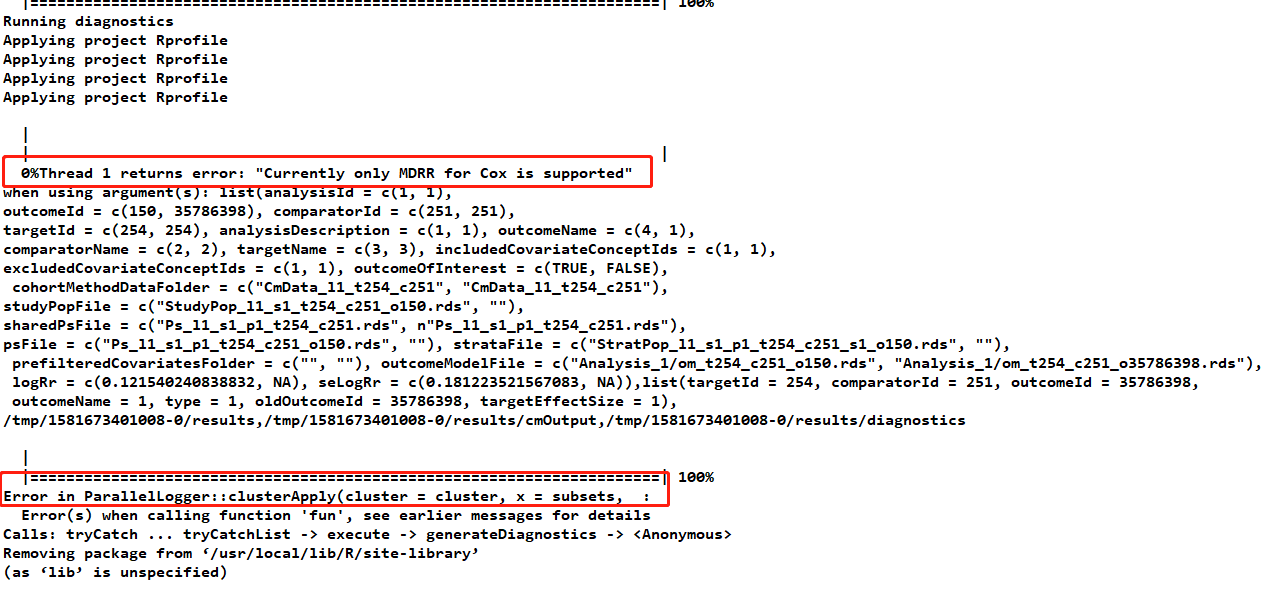
besides, then I use the atlas to do estimation and found there always are errors like the picture. I am wondering what is the mean of “currently only MDRR for cox is supported”, what is wrong? how can I solve this and the following error?
thank you very much@Eldar, yeah , i have a try about the drug_era, but the researcher want to constrain the drug exposure in the same one visit, while the drug_era hasn’t the samet visit choice.