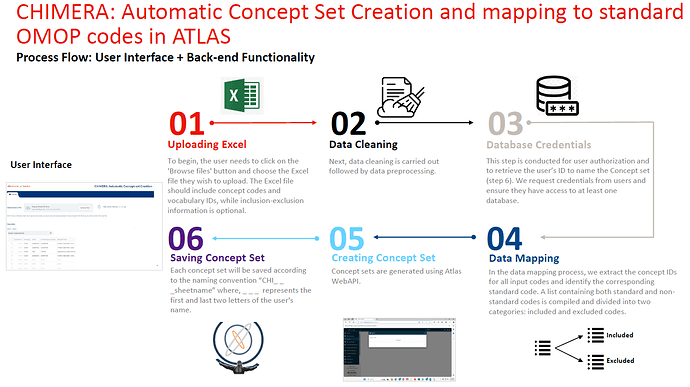
We are developing a tool to create and map concept sets automatically using Atlas. This will allow to scale the concept set creation and significantly reduce the time it takes. Two use cases prompted this development, 1) replicating concept sets from the literature 2) creating from scratch large concept sets or concept sets at scale (think on Phenotype Phebruary, hackathons, etc.).
The tool, called Chimera, is being tested internally (J&J) and we will be sharing it soon with the OHDSI community.
@Shahithya Lalitha has been key in the ideation and development. @aostropolets has been supporting us and we’ve learned a lot from the interactions with her. @Chris_Knoll has advised in solutions to use our system to host the tool, and @aandryc and Satyajit Pande are supporting with hosting the application in a platform/server so it’s fully functional.
Thus far, we can share the conceptual idea and next steps.
The first use-case was a real one when we were tasked with mapping the codes used to define systemic lupus erythematosus SLE according to several criteria: SLICC, ACR/EULAR.
There is a very thorough study by Walunas et al. in which the different set of criteria were translated to clinical codes in EHR/claims, they were also validated (Evaluation of structured data from electronic health records to identify clinical classification criteria attributes for systemic lupus erythematosus - PubMed).
The task to create these cohorts in Atlas was huge as more than 300 codes needed to be manually input into concept sets. This is why we decided to automatize as much as possible.
Eager to get the feedback from the OHDSI community as soon as we can share the Chimera tool!