I tried your code using the baseUrl that we use.
baseUrl <- `"https://atlas.astellasrwi.us/WebAPI"`
textToTest <- paste0("Testing update on ", as.character(Sys.time()))
originalDefinition <- ROhdsiWebApi::getCohortDefinition(cohortId = 2020, baseUrl = baseUrl)
modifiedCohortDefinition <- originalDefinition
modifiedCohortDefinition$description <- textToTest
ROhdsiWebApi::updateCohortDefinition(cohortDefinition = modifiedCohortDefinition, baseUrl = baseUrl)
ROhdsiWebApi::updateCohortDefinition(cohortDefinition = modifiedCohortDefinition, baseUrl = baseUrl)
Error in value[3L] :
Error with cohort definition: Error in .request():
! http error 403: Forbidden request.
You do not have permission to perform this action.
traceback()
7: stop(paste(“Error with”, category, “definition:”, error))
6: value[3L]
5: tryCatchOne(expr, names, parentenv, handlers[[1L]])
4: tryCatchList(expr, classes, parentenv, handlers)
3: tryCatch({
response ← .postJson(checkUrl, jsonExpression)
content ← httr::content(response)
}, error = function(error) {
stop(paste(“Error with”, category, “definition:”, error))
})
2: updateDefinition(baseUrl = baseUrl, definition = cohortDefinition,
category = “cohort”)
1: ROhdsiWebApi::updateCohortDefinition(cohortDefinition = modifiedCohortDefinition,
baseUrl = baseUrl)
I looked at the sourcecode for updateDefinition to follow where the error is occuring:
baseUrl <- gsub("/$", "", baseUrl)
arguments <- ROhdsiWebApi:::.getStandardCategories()
argument <- arguments %>% dplyr::filter(categoryStandard == 'cohort')
jsonExpression <- ROhdsiWebApi:::.toJSON(modifiedCohortDefinition$expression)
checkUrl <- paste(baseUrl, argument$categoryUrl, "check",
sep = "/")
Wondering if this checkUrl is correct ?
cat(checkUrl)
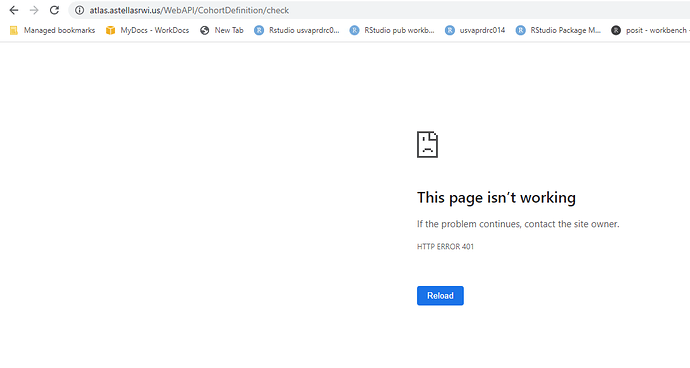
https://atlas.astellasrwi.us/WebAPI/cohortdefinition/check
response ← ROhdsiWebApi:::.postJson(checkUrl, jsonExpression)
Error in .request():
! http error 403: Forbidden request.
You do not have permission to perform this action.
Run rlang::last_trace() to see where the error occurred.
I am using latest version of ROhdsiWebApi 1.3.3 on R version 4.2.2.
I am unable to open the url link in the browser:
BaseUrl link does open in browser: https://atlas.astellasrwi.us/WebAPI/
Let me know if anything else is needed. I could not find the Atlas version we are using.