Works.  I am just very tired of my study-specific secret armbands. Very tedious to keep track of all our secret numbering systems. This would be a massive improvement.
I am just very tired of my study-specific secret armbands. Very tedious to keep track of all our secret numbering systems. This would be a massive improvement.
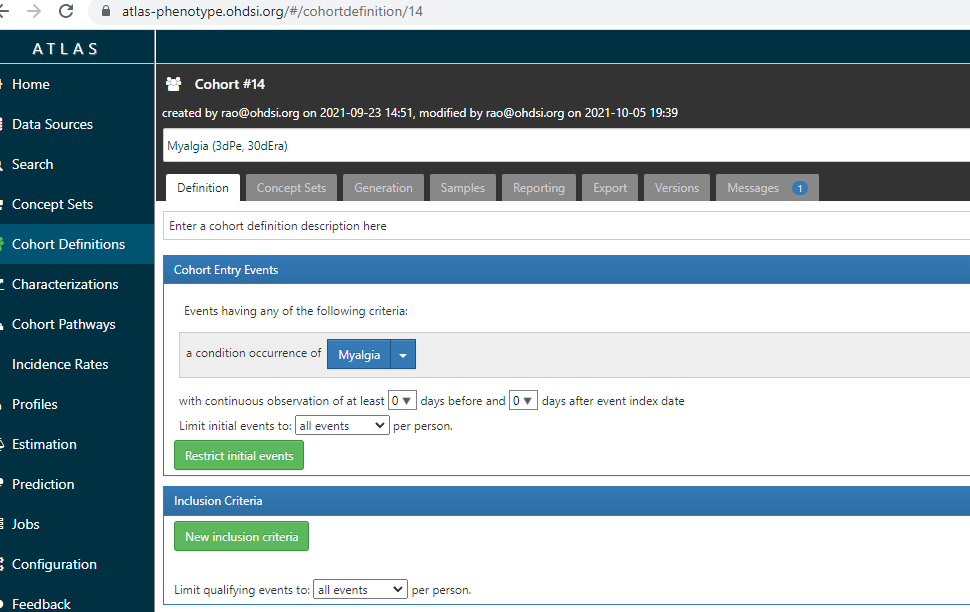
no secret numbers - all cohorts have the number that this atlas-phenotype.ohdsi.org gives. That cohort id will become immutable and be like a standard concept id in OMOP. So forever and ever till the end of OHDSI (which will never come) – the cohort id 14 will always be the same
For ever and ever - cohort id #14 will be Myalgia and this link will always give the same cohort definition
Thoughts?
I love the idea of an immutable and globally unique cohort identifier. What if we used a hash of the cohort json instead of or in addition to the ID in atlas-phenotype.ohdsi.org? That way if two hash IDs match you know for sure that the cohort definitions match.
Also I really like @Christian_Reich’s comments about conditions. My two cents is that there is no way around considering the data generating processes in rule-based phenotype algorithms. e.g. Logic to exclude rule out diagnoses does not make sense for registry data but does make sense for claims.
Thanks @Adam_Black
sure - do you have thoughts on a standard OHDSI way:
- maybe a standard hash algorithm that is used in all our software?
- an agreement on whether we do it on Cohort JSON or Cohort SQL (probably SQL as we may have cohort definitions that written in custom SQL not conformant to OHDSI Circe).
- An R function implementation of this - maybe in some HADES package like Cohort Generator?
The usable cohort definitions are maintained on ATLAS as shown here
I think in the short term simply using the ID in phenotype Atlas makes sense and is already implemented. In the longer term maybe we have URIs for concepts, concept sets, and cohort definitions.
@callahantiff - It seems like a cohort definition library fits nicely into the idea of the semantic web.
Do you think ideas in the semantic web community (e.g. How we identify things, URIs) could be applied here?
Hey @Adam_Black!
Very interesting observation and a very good use of the Semantic Web, maybe even more specific the FAIR (findable, accessible, interoperable, and reusable) data principles. I think I might need to learn more about what exactly the goals are? Are you trying to set-up a framework to make things more easily accessible and findable or are you strictly interested in a way to assign permanent resolvable URIs?
It’s also entirely possible that I am missing the larger goal altogether, so please feel free to let me know 
Based on the discussions in the community -
- ATLAS will be the official source of cohort definitions for the OHDSI Phenotype library (ready and available to community)
- The OHDSI Phenotype development and evaluation workgroup will be responsible to add/edit/remove cohort definitions to the atlas instance (ready and available to community)
- A Cohort Diagnostics study package will be maintained here https://github.com/OHDSI/PhenotypeLibrary/ which will be version controlled and referenceable. (ready and available to community)
- A shiny application with results will be posted on https://data.ohdsi.org/PhenotypeLibrary/ (to do)
Please see post here OHDSI Phenotype Library announcements