I need to use a set of ICD10 codes and DRG codes to define a cohort in Atlas.
Steps to replicate this bug:
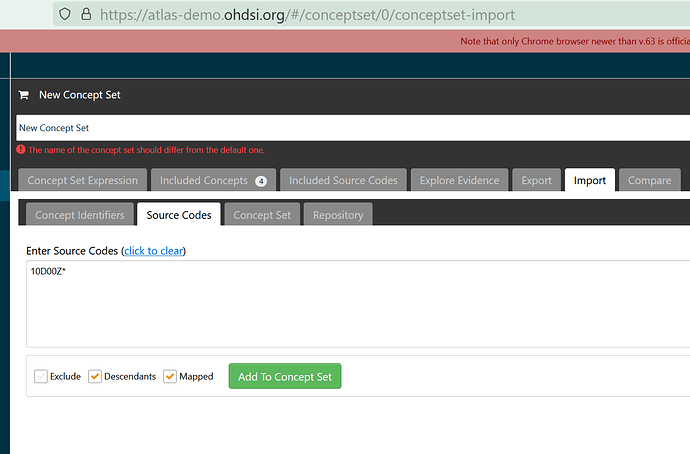
For example, certain partial matches work, such as for importing the source code “10D00Z*” in ATLAS
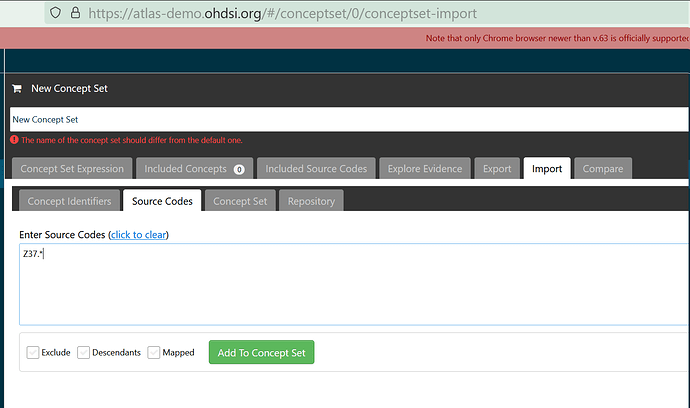
But certain others do not, such as “O00.*” or most others:
Is this inconsistency in Atlas documented anywhere? I am worried if I mix and match the asterisk operator I might be missing source codes during import, especially in case I might be using undocumented features.
To resolve this, I am manually going through the ICD database and creating lists for codes not supported by Atlas.
For example, I mapped the delivery hospitalization cohort in https://links.lww.com/AOG/C28 to the following set of codes:
# delivery codes
O80, O82
# delivery related group codes
765.00
765.01
765.02
765.03
765.04
765.05
765.06
765.07
765.08
765.09
766.0
766.1
766.2
766.21
766.22
767.0
767.1
767.11
767.19
767.2
767.3
767.4
767.5
767.6
767.7
767.8
767.9
768.0
768.1
768.2
768.3
768.4
768.5
768.6
768.7
768.70
768.71
768.72
768.73
768.9
774.0
774.1
774.2
774.3
774.30
774.31
774.39
774.4
774.5
774.6
774.7
775.0
775.1
775.2
775.3
775.4
775.5
775.6
775.7
775.8
775.81
775.89
775.9
# cesarean/delivery codes
10D00Z*
10D07Z*
10E0XZZ
# outcome of delivery
Z37.0, Z37.1, Z37.2, Z37.3, Z37.4
Z37.50, Z37.51, Z37.52, Z37.53, Z37.54
Z37.59
Z37.60, Z37.61, Z37.62, Z37.63, Z37.64, Z37.69, Z37.7, Z37.9
# exclude: abortion outcome
O00.0
O00.1
O00.2
O00.8
O00.9
O01.0
O01.1
O01.9
O02.0
O02.1
O02.8
O02.81
O02.89
O02.9
O04.5
O04.6
O04.7
O04.8
O04.80
O04.81
O04.82
O04.83
O04.84
O04.85
O04.86
O04.87
O04.88
O04.89
O07.0
O07.1
O07.2
O07.3
O07.30
O07.31
O07.32
O07.33
O07.34
O07.35
O07.36
O07.37
O07.38
O07.39
O07.4
O08.0
O08.1
O08.2
O08.3
O08.4
O08.5
O08.6
O08.7
O08.8
O08.81
O08.82
O08.83
O08.89
O08.9
So sorry if this is covered elsewhere, it’s my first time trying to replicate this type of study using Atlas.
Thanks so much!
Jaan