@schuemie, hitting a bit of an issue with the very last step of a PLE study.
I’m unable to package the results into a Zip. What package executes this function? Maybe I’m missing it.
I’m getting this as my error:
|Packaging results Exporting analyses - cohort_method_analysis table - covariate_analysis table Exporting exposures - exposure_of_interest table Exporting outcomes - outcome_of_interest table - negative_control_outcome table Exporting metadata - database table - exposure_summary table - comparison_summary table - attrition table |===============================================================================================| 100% - covariate table - cm_follow_up_dist table |===============================================================================================| 100% Exporting main results - cohort_method_result table Performing empirical calibration on main effects |===============================================================================================| 100% censoring 0 values (0%) from targetSubjects because value below minimum censoring 0 values (0%) from comparatorSubjects because value below minimum censoring 0 values (0%) from targetOutcomes because value below minimum censoring 0 values (0%) from comparatorOutcomes because value below minimum - cm_interaction_result table |===============================================================================================| 100% Exporting diagnostics - covariate_balance table |===============================================================================================| 100% - preference_score_dist table |===============================================================================================| 100% - propensity_model table |===============================================================================================| 100% - kaplan_meier_dist table Computing KM curves |===============================================================================================| 100% Writing to single csv file
Adding results to zip file
Error in .jcall(“RJavaTools”, “Ljava/lang/Object;”, “invokeMethod”, cl, : java.lang.RuntimeException: java.io.FileNotFoundException: ./IUD/AmbEMR/export/ResultsAmbEMR.zip (No such file or directory) Warning: Error ‘cannot open the connection’ when writing log to file './IUD/AmbEMR/log.txt. Removing file appender from logger.|
It can’t open the results because it is not able to execute writing the results zip. All the other csvs are written. Any thoughts on how to debug this?
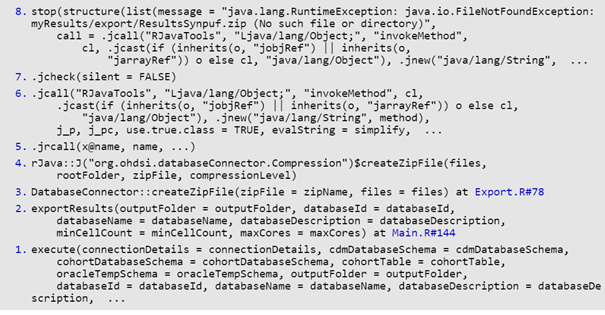
traceback()
7: stop(list(message = “java.lang.RuntimeException: java.io.FileNotFoundException: ./IUD/AmbEMR/export/ResultsAmbEMR.zip (No such file or directory)”,
call = .jcall(“RJavaTools”, “Ljava/lang/Object;”, “invokeMethod”,
cl, .jcast(if (inherits(o, “jobjRef”) || inherits(o,
“jarrayRef”)) o else cl, “java/lang/Object”), .jnew(“java/lang/String”,
method), j_p, j_pc, use.true.class = TRUE, evalString = simplify,
evalArray = FALSE), jobj = new(“jobjRef”, jobj = <pointer: 0x6bf5750>,
jclass = “java/lang/RuntimeException”)))
6: .jcheck(silent = FALSE)
5: .jcall(“RJavaTools”, “Ljava/lang/Object;”, “invokeMethod”, cl,
.jcast(if (inherits(o, “jobjRef”) || inherits(o, “jarrayRef”)) o else cl,
“java/lang/Object”), .jnew(“java/lang/String”, method),
j_p, j_pc, use.true.class = TRUE, evalString = simplify,
evalArray = FALSE)
4: .jrcall(x@name, name, …)
3: rJava::J(“org.ohdsi.databaseConnector.Compression”)$createZipFile(files,
rootFolder, zipFile, compressionLevel)
2: DatabaseConnector::createZipFile(zipFile = zipName, files = files) at Export.R#78
1: exportResults(outputFolder, databaseId, databaseName, databaseDescription,
minCellCount = 5, maxCores)