We did! We’ve got some information posted at: https://github.com/NUSCRIPT/OMOP_to_Graph and are working on a paper now.
We created a 3NF version of the OMOP CDM for a Bristol-Myers Squibb PoC. We used views on the CDM to recreate OMOP CDM tables. Recently we built a Neo4j implementation of OMOP to build an OMOP Knowledge Graph. The added value of a graph DB in this case is for hypothesis generation. You can start with any concept in the CDM vocabulary and follow the relationships (shared attributes) to discover relationships that would not find in a RDBMS without knowing what you are looking for or writing an expensive query. The interesting relationship just pops out to SMEs.
I have created a graph version of the OMOP Standard Vocabulary. If anyone is interested please let me know.
Hi Ed, I’m working on a related project and would love to learn more about how you constructed your graph and use it as a reference. Are the relationship/source files available?
Hi, that’s great! I’m interested on this!
Hi @lrasmussen, I just recently got interested in the topic of combining OMOP/neo4j. Are you still working on the paper or is it maybe even already published?
Hi @eacker, I’d also be super interested to find out more! Are you sharing this anywhere?
Hi Carina,
I am looking for data to try loading the database.
Hi @eacker ,
I am interested to learn more about your experience as I am also working on the same task. Happy to discuss more my approach with you
Thank you
@Adam_Bouras @eacker @Carina_D
I would be interested in collaborating. I have been implementing some vector databases for NLP, but previously I have used Neo4j for this and storing vectors there for clinical notes.
I think adapting the CDM to other databases will be important going forward with some of the applications we may want to implement. Relational is great, but it just doesn’t work in some cases.
Same here! We work on a OMOP → KG mapping for clinical studies at the German Center for Diabetes Research (DZD). Should we set up a short call to see where our projects overlap?
To be honest, I don’t have any concrete project right now but I’m curious about the topic, would like to find out how this could be applied in OMOP data and especially what new analysis opportunities are enabled with this. I’d be happy to have a call to explore this!
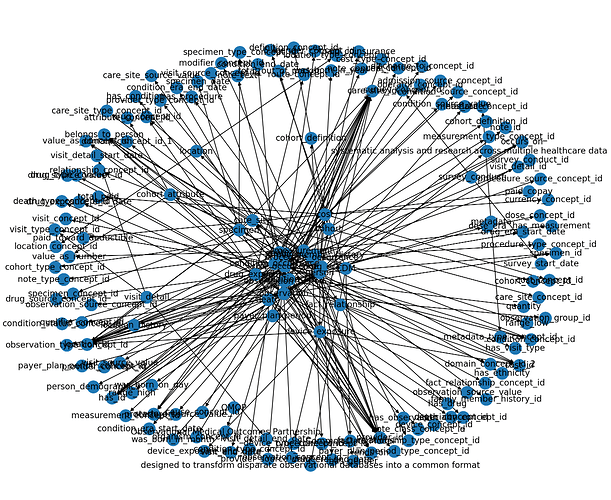
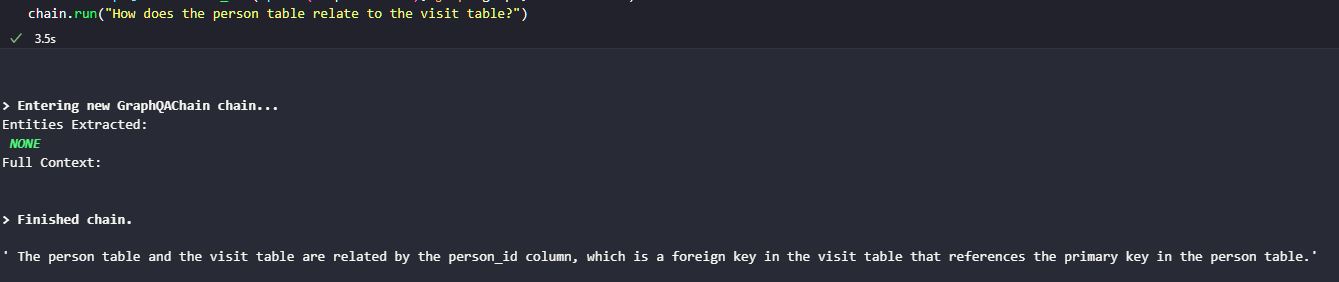
Generating a Digraph wasn’t too bad, and it is quiet useful to work out some of the issues with large language models understanding the CDM
If you’d like to contribute in making tuples, the CSV I use to generate the nodes can be found here.