@Gowtham_Rao @Patrick_Ryan @Azza_Shoaibi is there a Phenotype Phebruary meeting today (Fri Feb 10, 2023)? What time EST/CST? I see a Teams link above, but its not on my calendar so have to check my availability.
Yes, we’re meeting today at 9 am EST (very soon). I am afraid I don’t know how to obtain the link… @Gowtham_Rao should know how.
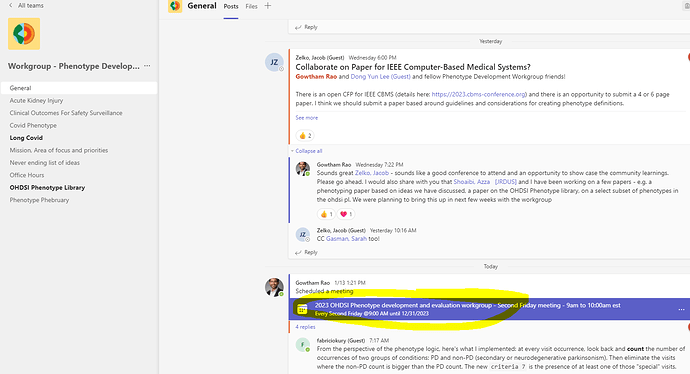
@allanwu Today we have the standing meeting of the OHDSI Phenotype Development and Evaluation Workgroup
To access meeting information - please use the OHDSI MS teams as shown in screenshot
Proposed agenda
Sorry I have a conflict at that time; I will see if I can join at the end. I see the Phea discussion coming up near the end; I’ll reply to @fabkury logic of the Phea SQL addition to tiered consensus in that thread to contribute.
Hi, are there instructions anywhere for how to do this? I have executed CohortDiagnostics and got results but I do not know how to make them available on data.ohdsi.org. Thanks!
Great question.
If the data object is <100mb and you want to make a shiny app on data.ohdsi.org, then the steps are:
- Go to the Shiny repo: GitHub - OHDSI/ShinyDeploy: Shiny apps in this repository will be automatically deployed to the OHDSI Shiny server.
- Log in to your github account
- Create a fork:
- Select your user account name as the place to create the fork
- Clone your fork
- Create a folder in your clone for your shiny, e.g. “SLECohortDiagnostics”
- [Important] Download the latest version of CohortDiagnostics as zip from GitHub - OHDSI/CohortDiagnostics: An R package for performing various cohort diagnostics. .
1.Unzip and find the folder CohortDiagnostics\inst\shiny\DiagnosticsExplorer folder - Copy all files, as is, from the folder above to the folder you created in Step 6
- Create a new folder in your folder (created in step 6) – rename it as “data”
- Add your .sqlite file to the ‘data’ folder
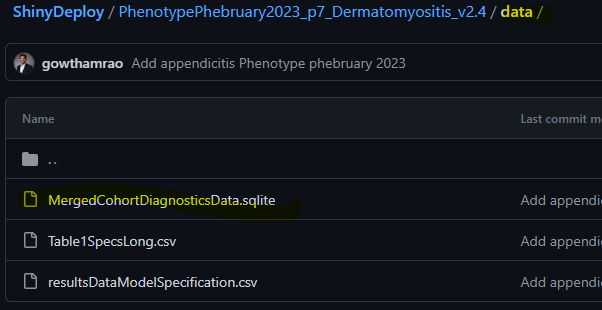
- Test your app: Click on the below file object in your folder to open Rstudio and run app locally by clicking on RunApp after opening server.R file.
- Commit your work to github
- Create a Pull Request
a. Click
b. Click
- Enter the information for your pull request and save
- Notify a repo administrator (erica, Gowtham Patrick, Jenna, Martijn) of your pull request
- Will be available at https://data.ohdsi.org/SLECohortDiagnostics
If your .sqlite file > 100mb, then you cannot host it on github because of file size limitation - you will have to load to a postgres server exposed to the internet and data.ohdsi.org shiny server should know how to talk to it. Thats more complicated.
Thank you, really appreciate this. Will work on these steps.