I don’t know what you mean by this: clinical drugs roll up to ingredients via the concept_ancestor hierarchy.
Hi all, I have a propose to add a new concept class for LOINC attributes - “GENE”, we can find link between measurement and this attribute in
Some examples:
| loincnumber | longcommonname | partnumber | partname | parttypename |
|---|---|---|---|---|
| 44606-2 | ALOX3+ALOX12B gene mutations found [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP288622-6 | ALOX3 gene | GENE |
| 44606-2 | ALOX3+ALOX12B gene mutations found [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP288621-8 | ALOX12B gene | GENE |
| 42712-0 | AML/MDS gene 7q31 deletion [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP288623-4 | AML gene | GENE |
| 42712-0 | AML/MDS gene 7q31 deletion [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP288634-1 | MDS gene | GENE |
| If we do this, it will become easier for researchers to work with genomic data. | ||||
| @Vojtech_Huser @Christian_Reich @Dymshyts @Polina_Talapova @Alexdavv @zhuk What do you think about this? |
It would. We may not call it “GENE” though. More like “Genetic aberration”
@Denys_Kaduk, with a new class ‘Gene/Genetic aberration’ you will get just a full set of associated genes for each test. You won’t even know whether it’s searching for an alteration/mutation or just gene detection analysis. Also, you cannot distinguish between single genes and gene combinations (just 2 links are created for the latter).
For now, we’re planning to have the following:
| loincnumber | longcommonname | partnumber | partname | relationship_id |
|---|---|---|---|---|
| 44606-2 | ALOX3+ALOX12B gene mutations found [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP227586-7 | ALOX3+ALOX12B gene targeted mutation analysis | Has component |
| 42712-0 | AML/MDS gene 7q31 deletion [Identifier] in Blood or Tissue by Molecular genetics method Nominal | LP227622-0 | AML+MDS gene 7q31 deletion | Has component |
As a Component, you have a gene/gene combination + type of analysis in a single attribute concept. Unfortunately, LOINC doesn’t use ‘targeted mutation analysis’/ ‘deletion detection’, etc. as precise ‘Molgen’ METHODs, so you need to sort out all these components in any kind of genomic analysis anyway.
Also, we are thinking about adding the hierarchical links between test COMPONENTs and more general COMPONENTs derived from ‘DetailedModel’ and ‘SyntaxEnhancement’ model (what you probably need as Genes).
| partnumber | partname | relationship_id | partnumber | partname |
|---|---|---|---|---|
| LP227586-7 | ALOX3+ALOX12B gene targeted mutation analysis | Is a | LP36747-1 | ALOX3+ALOX12B gene |
| LP227622-0 | AML+MDS gene 7q31 deletion | Is a | LP36196-1 | AML+MDS gene 7q31 |
@Vojtech_Huser @Christian_Reich @rimma @Denys_Kaduk What option is preferable? Or we can implement both.
We need one more community advice for Loinc Parts incorporation into CDM for the upcoming LOINC release.
Should we make them Standard?
There are a few options:
- Make them standard. LOINC Parts can be helpful in the mapping process but are not extremely necessary. There are a lot of possibilities to perform mapping to SNOMED or other vocabularies without mentioning LOINC Parts.
- Make them non-standard.
- The best option would be to make it all classification concepts so users can query concept ancestor to get Lab tests with any specified method, scale, property, component, etc. as our initial plan was. BUT since we’ve decided to use the same relationships for SNOMED and LOINC, making parts ‘C’ concepts will require complete SNOMED rearrangement and have a lot of consequences. That’s why not the best decision, unfortunately.
@Christian_Reich @Vojtech_Huser what do you think?
Agree with 3
Impossible solution is always the best solutions 
SNOMED attributes are Standard concepts so how can they be ancestors of SNOMED tests?
Relationships between tests and attributes become vertical hierarchical, right?
Otherwise, we need to split back SNOMED and LOINC relationships for attributes and build different logic for SNOMED and LOINC.
Question about this approach: using a concept ancestor using for a higher level class of measurement may lead to the lower-level measurement concepts being selected, but you may get different units for those different children. So, what would a query look like where you search on a parent concept, but then need to find the appropriate unit/measure for the possible children?
Ad making parts standard: I don’t know enough about all the rules Athena team is using when doing changes to the vocabulary. And also which LPs were even now made into concepts and which were not. I was looking at LOINC in order to map HIV viral load data. E.g., at methodless codes in LOINC and how often there are such codes. (if interested, my ugly code is here project/lab/loinc-parts-v002.Rmd at master · vojtechhuser/project · GitHub )
I looked at result of this current query
http://athena.ohdsi.org/search-terms/terms?vocabulary=LOINC&conceptClass=LOINC+Hierarchy&standardConcept=Non-standard&page=1&pageSize=15&query=
(but no major conclusions from reviewing it)
One bonus remark
e.g., LP for methods should probably not be standard. e.g. LP6464-4 | Probe.amp.tar
and currently it is not even a concept. (this should become one but not a standard one)
(the LP example is taken from https://raw.githubusercontent.com/vojtechhuser/project/master/lab/LoincPartLinkPrimary.csv )
at LOINC upcoming conference, I see the following on agenda
Automated hierarchical crosswalk between LOINC Laboratory tests and SNOMED CT Measurements
Photo forthcomingPhoto forthcomingPolina Talapova, MD
Odysseus Data Services
Eduard Korchmar
Odysseus Data Services
It is known that the purpose of the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) is to standardize the representation format of healthcare data. It has been adopted by the Health Data Sciences and Informatics (OHDSI) collaborative, a multi-stakeholder, to create open-source solutions that bring out the value of observational health data through large-scale analytics.In the OMOP CDM, such ontologies as SNOMED CT and LOINC are used as Gold standards for the transformation of raw Observations and Measurements into the standardizes ones. Moreover, Regenstrief Institute and SNOMED International have formed a long-term collaborative relationship with the objective of developing coded content to support order entry and result reporting.
Material and methods: the LOINC-SNOMED collaborative has provided a mapping from LOINC Terms to SNOMED pre-coordinated concepts. Using this, we have built a hierarchical crosswalk that embeds more granular LOINC Laboratory tests into the SNOMED CT Hierarchy of Measurements. The approach is based on both sophisticated logic and an automatic match of common attribute combinations via SQL. Hierarchical links are represented by “Is a” relationship that implies that the LOINC Term semantically resides below the SNOMED CT concept.
Results: We have obtained more than 12,000 hierarchical links from the LOINC Laboratory Tests to SNOMED Measurements. This approach gives the opportunity to reproduce the automatic mapping with respect to every LOINC update.
@Polina_Talapova - can you comment on if those will be released (or have been released) in OMOP Vocab ?
I understand that the mapping the abstract references was done few years ago (and not sure if there is commitment to keep it updated) and newest LOINC terms may not have this mapping. (so OHDSI would have to “maintain it”)
@Vojtech_Huser, the hierarchical mapping has not released yet (but I am looking forward to this).
I plan to map more LOINC Parts to SNOMED attributes. This will help to build more links between LOINC Laboratory tests (including new Terms) and SNOMED Measurements.
Curious: How are you going to deal with the many-to-many valid mapping outputs that LOINC generates?
Since we do not neccessary look for equal mappings in all cases and OMOP CDM allows for multiaxial hierarchy, our main concern is that all LOINC concepts become descendants of all SNOMED concepts they should be descendants of. It is absolutely expected that LOINC concepts will have multiple SNOMED parents and that some SNOMED concepts may have multiple LOINC descendants.
Also of note is this call by LOINC (effort to update top2000 tests). Most frequently used LOINC codes – LOINC
In addition, LOINC effort for groups is relevant and was mentioned in conversations I had with various folks.
How will you handle the multiple valid mappings that LOINC generates within LOINC itself that are not interoperable? LOINC urgently needs high level terms.
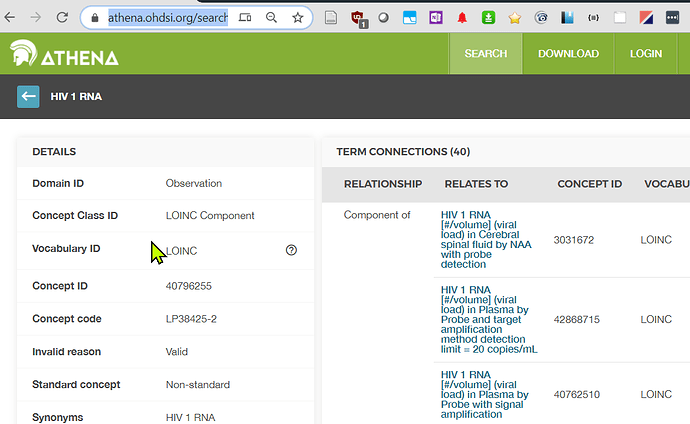
I am pleased to see that as of May 2020 (and probably earlier) - OMOP Vocab now has implemented the Loinc Part inclusion as I suggested.
See example here
This component is now an Athena concept
Athena for ‘HIV1 RNA’
and has relationships such as ‘component of’