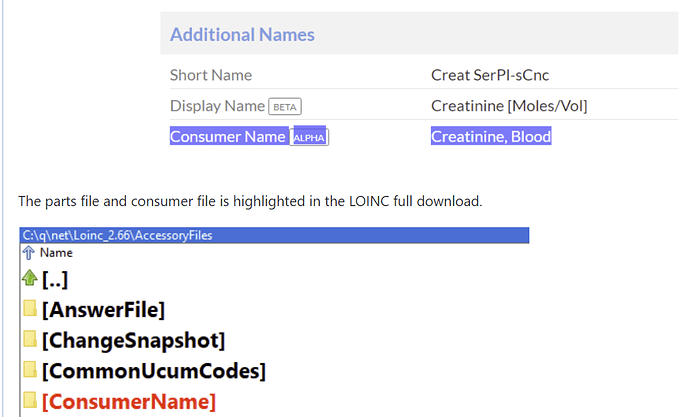
@Denys_Kaduk, with a new class ‘Gene/Genetic aberration’ you will get just a full set of associated genes for each test. You won’t even know whether it’s searching for an alteration/mutation or just gene detection analysis. Also, you cannot distinguish between single genes and gene combinations (just 2 links are created for the latter).
For now, we’re planning to have the following:
| loincnumber |
longcommonname |
partnumber |
partname |
relationship_id |
| 44606-2 |
ALOX3+ALOX12B gene mutations found [Identifier] in Blood or Tissue by Molecular genetics method Nominal |
LP227586-7 |
ALOX3+ALOX12B gene targeted mutation analysis |
Has component |
| 42712-0 |
AML/MDS gene 7q31 deletion [Identifier] in Blood or Tissue by Molecular genetics method Nominal |
LP227622-0 |
AML+MDS gene 7q31 deletion |
Has component |
As a Component, you have a gene/gene combination + type of analysis in a single attribute concept. Unfortunately, LOINC doesn’t use ‘targeted mutation analysis’/ ‘deletion detection’, etc. as precise ‘Molgen’ METHODs, so you need to sort out all these components in any kind of genomic analysis anyway.
Also, we are thinking about adding the hierarchical links between test COMPONENTs and more general COMPONENTs derived from ‘DetailedModel’ and ‘SyntaxEnhancement’ model (what you probably need as Genes).
| partnumber |
partname |
relationship_id |
partnumber |
partname |
| LP227586-7 |
ALOX3+ALOX12B gene targeted mutation analysis |
Is a |
LP36747-1 |
ALOX3+ALOX12B gene |
| LP227622-0 |
AML+MDS gene 7q31 deletion |
Is a |
LP36196-1 |
AML+MDS gene 7q31 |
@Vojtech_Huser @Christian_Reich @rimma @Denys_Kaduk What option is preferable? Or we can implement both.