Hi Martijn,
I ran it using version 2.1.5 of FeatureExtraction.
@JUNGEUN_PARK: which versions of CohortMethod and FeatureExtraction are you running?
@schuemie
I was running vesion 2.1.3 of FeatureExtraction and just updating it solved my problem!
Thank you very much for your help. 
I wonder why I keep getting this error : 00904 Invalid with DibaetesTxPackage
Below is the error report.
Any advice would be appreciated. Thank you.
DBMS:
oracle
Error:
java.sql.SQLSyntaxErrorException: ORA-00904: : Invalid Identifier
SQL:
CREATE TABLE gi05a2z4ohdsiT2DstudyPop (rowId FLOAT,subjectId FLOAT,treatment FLOAT,cohortStartDate DATE,daysFromObsStart FLOAT,daysToCohortEnd FLOAT,daysToObsEnd FLOAT,outcomeCount FLOAT,timeAtRisk FLOAT,daysToEvent FLOAT,survivalTime FLOAT)
R version:
R version 3.4.2 (2017-09-28)
Platform:
x86_64-w64-mingw32
Attached base packages:
- stats
- graphics
- grDevices
- utils
- datasets
- methods
- base
Other attached packages:
- gridExtra (2.3)
- DiabetesTxPath (0.1.0)
- EmpiricalCalibration (1.3.6)
- CohortMethod (2.6.2)
- FeatureExtraction (2.1.5)
- DatabaseConnector (2.0.2)
- SqlRender (1.4.6)
- OhdsiSharing (0.1.3)
- OhdsiRTools (1.5.3)
- Cyclops (1.3.0)
- rJava (0.9-9)
I’m pretty sure it’s because ROWID is a reserved word in Oracle. @rohitv: any idea where this SQL originated from?
Ah, wait, I see it. This call to insertTable would do the trick. @rohitv: you may want to change the column names of studyPop before you send it to the server.
I usually use
colnames(studyPop) <- SqlRender::camelCaseToSnakeCase(colnames(studyPop))
to get snake_case field names, which is our standard for SQL.
Hi Martijn,
Updated.
Regards
Rohit
Thank you. I thought that the rowID might be causing the error, but had no idea how to fix that.
Still just waiting for any solutions.
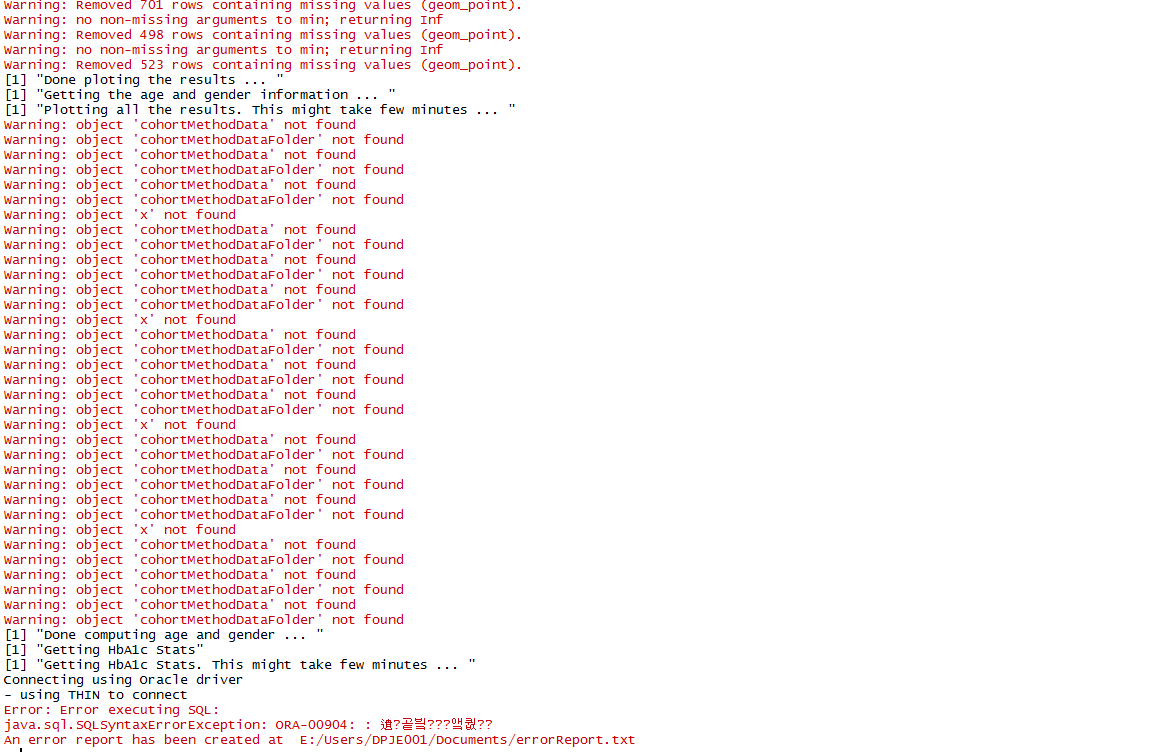
Also, there was an error "'CohortMethodData' not found"
I wonder if this error is also related to Oracle.
@JUNGEUN_PARK Could you update the packages (especially, SqlRender like below) and try again?
devtools::install_github("ohdsi/SqlRender")
@SCYou
Tried but didn’t really work. Thank you for the idea ,though 
Again, just guessing, but I think the problem with the cohortMethodData is caused here. The loadCohortMethodData function returns a cohortMethodData object, but here it is not assigned anywhere. The subsequent calls to cohortMethodData will throw errors. Probably that line should read:
cohortMethodData <- loadCohortMethodData(paste(results_path,"deleteMeBeforeSharing/CmData_l1_t1_c2",sep=""))
@rohitv: note that file.path is preferred over paste when creating a path.
I tried, but still didn’t work.
And still, cohortMethodData not Found error comes up.
I appreciate your suggestions. 
Hi @JUNGEUN_PARK. What exactly did you try? Do you have a new version of the study code I can inspect?
Hi @schuemie,
I tried the following line.
cohortMethodData <- loadCohortMethodData(paste(results_path,"deleteMeBeforeSharing/CmData_l1_t1_c2",sep=""))
I had to change more words to make this package work, and now (at leaste I think) I get all the outputs as expected.
But I still get 'CohortMethod' Data not found error. Maybe it is because of the data or the result of analysis?!
Thank you as always.
- I have one more question. Everytime I run this, it gives me slightly different(or sometimes quite a lot) results. Is it because I set the maxcores=1? What is the reason of the different results?
If i search for ‘cohortMethodData’ in the same file I see many occurrences of loadCohortMethodData, without assigning the output to a variable. Did you fix all of them?
@rohitv: perhaps it would also be a good idea to fix the original study code?
I’m not very familiar with this code (I didn’t write it), but CohortMethod itself (which I did write) can give different answers on different runs because the sampling for the cross-validation for Cyclops (to establish the optimal hyper-parameter) is random and can differ between runs. In order to make things 100% deterministic, you can try setting the random seed in the Cyclops control object when calling createPs.
Yes, I fixed all of them and still had the same error above.
Thank you. 
Hello,
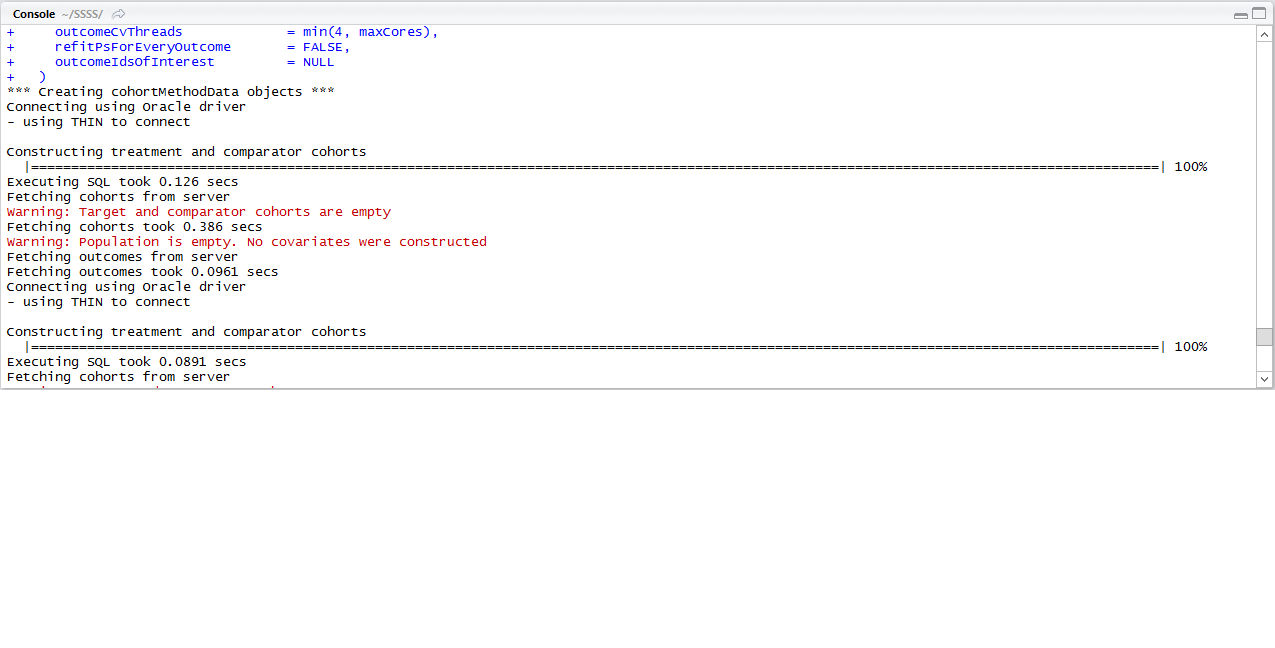
I’m trying to run runcmAnlaysis() but keep getting Target and Comparator cohorts are empty error.
I think I have done pretty much what others do.
I created exposure table and outcome table to my schema for multiple anlayses,
and there certainly are the tables I created in the schema. (I could check it with oracle)
In the outcome table, cohort_definition_id is c(1,2)
In the exposure table, cohort_definition_id is c(3,4,5,6,7,8,9,10)
and c(3,5,7,9) for target, c(4,6,8,10) for comparator cohort.
I also created drugComparatoroutcomesList for this.
and I used table_cohort_id and table_cohort_id argument in loadRenderTranslateSql().
And (at least I think) there should be no error.
I’ll edit with some more details if necessary. Thank you!
If you open the cohortMethodData object generated by the code you can call getAttritionTable() to see why there’s no-one left.
Thank you very much! @schuemie
Now I’m certainly sure I’m lost here.
Running getAttritionTable(cohortMethodData),
- original cohorts had 1,621 patients, and 2.Removed subs in both cohorts had 0 patients.
I’ve ran this exactly same analysis before so I know I’m doing something wrong.
Maybe the exposureTable and outcomeTable is the problem.
but I set the exposureDatabaseSchema and outcomeDatabaseSchema to the same schema I’m using. and named both tables of exposure and outcome “ohdsi_Neph” and “ohdsi_Neph_outcome”.
and in Orcale, I could see those tables exist with
select * from myschema.ohdsi_Neph
and, the values of COHORT_DEFINITION_IDs are exactly same as the targetcohortID or comparatorcohortID.
(for example, I set targetcohortID c(3,5,7,9) and comparatorcohortID c(4,6,8,10))
Any advice or tips would be so much helpful. Thank you.
Below is part of the R code I used to create target and comparator cohorts.
sql <- SqlRender::loadRenderTranslateSql(sqlFilename=sqlFilename, packageName= packageName, dbms=attr(connection,"dbms"), cdm_database_schema=cdmDatabaseschema, target_datbase_schema=targetDatabaseschema, target_cohort_table=targetCohortTable, target_cohort_id=1)
DatabaseConnector::executeSql(connection=connection, sql=sql)
Hi @JUNGEUN_PARK. So it seems there are indeed people in the exposureTable, but the cohorts completely overlap, and you’ve specified for people that appear in both target and comparator cohort to be removed, so no one remains.
When calling createTargetComparatorOutcomes(), do you set targetId and comparatorId to different values? Here’s an example of how to use createTargetComparatorOutcomes.