There should not be any human concepts in the veterinary extension from Virginia Tech. The moduleId should be 332351000009108 = Veterinary Terminology Services Laboratory maintained module (core metadata concept) for all the concepts. If there is a discrepancy, please let me know.
I believe the original request to add the Veterinary Extension was sent by Chris Brandt. Just want to make sure you have the correct files. The Veterinary Extension files can be downloaded at:
Yes,
and yes, we are working on this one
The thing is Veterinary Extension can work as a self-contained vocabulary, so
there are concepts that help to make SNOMED structure, i.e. Attributes (70258002 Ankle joint structure), general concepts that works for human and animals (77386006 Pregnant, 91302008 Sepsis etc).
Also some veterinary terms already exist in Human SNOMED, for example
Organisms: 61839006 Cornish chicken, even viruses that affects animals (71448008 Canine minute virus) etc.
Attached file contains all such a concepts:
common_concepts.xlsx (132.0 KB)
Thanks Dmytry,
The problem with creating a veterinary vocabulary with only the veterinary extension is that many human concepts are used to code animal diseases without any concepts from the veterinary extension. I thought the vocabulary you were creating was just the concepts, relationships and descriptions in the Veterinary Extension. I think a freestanding vocabulary is a great idea. Since VMDB has 15 years and approximately 1.7 million visits codes, I can query the database and provide all the human concepts that have been used for coding from our submissions. Those concepts plus the veterinary extension will provide a self-contained vocabulary and allow for comparing animal and human diseases. That vocabulary should met the needs of anyone interested in coding / transforming animal records. Let me know if that will be helpful.
Problem with this approach is that there will be two concepts for Sepsis, one from SNOMED and another one from SNOMED Vet. They really mean the exact same thing. One of our principles is to have only one standard concept per semantic entity per Domain, and the other concepts map to it. If we did it as you suggest we would go into the opposite direction and violate that principle.
The alternative is this: Veterinarians have to use the union of SNOMED and SNOMED Vet for their work. The rest of the community, which is the vast majority, can continue just with SNOMED as if nothing happened. Similar to the US researchers use RxNorm for drugs, and everybody outside has to say where vocabulary_id in ('RxNorm', 'RxNorm Extension') to make use of that combined universe. Works well.
The only downside to that approach is that your colleagues will have certain non-veterinary concepts to deal with, like “Bipolar Disease”. Can they live with that?
From a SQL, programming perspective either approach is fine. As was said - using “vocabulary_id in (‘RxNorm’,‘RxNorm Extension’)” construct is common. For how folks are using and constructing SQL now, I think one concept principal is the preferred method. It also simplifies the decision logic of choosing which vocabulary to to map the example “Sepsis” to and maintaining the consistency if the query is run on both the human and animal sides. This also impacts on the application side as well - displays one concept choice instead of two and have to display which vocabulary the concept is attached - more screen space required.
Using a union of SNOMED and SNOMED Vet is perfectly acceptable. In fact, that is how VMDB incorporates SNOMED and the veterinary extension. Just to state the obvious since I am a bit of a newbie to OHDSI, to use the CDM, veterinarians would use both the SNOMED vocabulary and the SNOMED veterinary extension vocabulary. If the veterinary vocabulary contains only those with the moduleId for the veterinary extension, then there will not be any duplicate concepts and fits the principles for OHDSI? Is that stated correctly?
Thanks again.
Exactly. Thanks guys.
We found a typo:
there’s concept_code =‘89001000009106’ with description = ‘Cracticus mentalis (oranism)’
- oranism instead of organism.
Where to report it?
Hi, not knowing the context, here’s what I did to come up with my below suggestion.
89001000009106, clearly a SNOMED-code.
But not in international release.
Looked at 1000009 here: https://cis.ihtsdotools.org/info/index.html?home=namespaces
This shows it’s VetMed.
Had downloaded that extension, and saw the error was there too (so not a copy-paste error).
So either you approach info@snomed.org, as mentioned on the site mentioned earlier.
Or, contact the owner mentioned in previous versions of the list found here: https://confluence.ihtsdotools.org/download/attachments/6160816/SNOMED_CT_Namespace_Registry%20-%20OFFICIAL%2020141021.pdf?version=1&modificationDate=1426061583000&api=v2&download=true
That gives the answer:
Email: jwilcke@vt.edu
Best, Ronald
Thanks @ronaldcornet.
I like really like your logic, you act like a detective here:)
We found a bunch of other innaccurracies, so we’ll report them.
I’m happy to inform that Veterinary Extension is available for download in Athena.
Vocabulary_id =‘SNOMED Veterinary’
This is great! Thanks for getting the vocabulary setup.
Just want to add my thanks for creating that.
You’re welcome:)
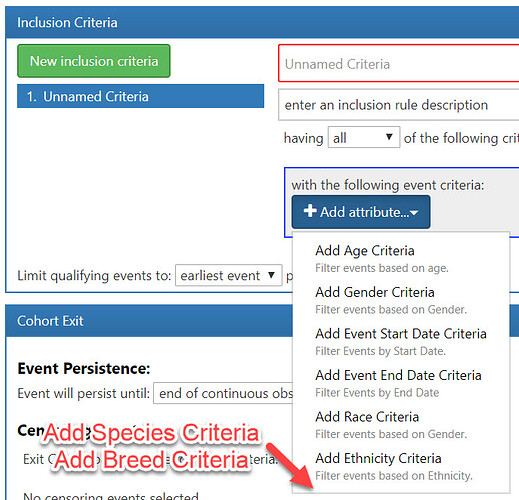
With the Veterinary extension in place (thanks Dmytry!), can we look again at the structure for storing Species and Breed?
Wayde originally suggested 2 options:
- Use existing Person table and add Species and Breed columns which would include OHDSI concept IDs (from SNOMED / Vet Extension)
- Rename Person tables/fields to represent Animal. Create additional tables to represent owner and link with many-many relationships
Michael Kahn proposed:
3) Leave OMOP tables/columns “as is” with their intended semantics and add satellite table(s) linked to the standard OMOP tables for all animal-specific attributes.
I agree with Manlik’s comment:
“Utilizing Observations to hold species and breed is certainly one viable approach, but species and breed are at level of the patient’s demographics along with age and sex.” So I feel like option 2 provides a simple way to add this critical data for veterinary use. Yes, animal data would be housed in a table labeled ‘Person’, but I think if we’re going to be using Atlas and the other tools for Veterinary use, we’ll need to be flexible in the labels for things. I’m not expecting the icon next to ‘Cohort Definitions’ to change from 3 people to a picture of a dog, cat, and horse ![]()
Let’s assume (for a moment) that we were able to convince the community that Option 2 was the right way to go… We then need to look at modifying the code for Atlas to use that data appropriately. One of my developers has looked at this and found adding Breed and Species to the initial Demographic Picker was relatively easy. Well – easy to ADD it to the list… we still need to wire it up so that it knows how to store the cohort definition and how to construct appropriate queries to return results for the cohort.
It seems that having a very similar structure to race/ethnicity criteria would allow this feature to be added more easily than if it was a completely separate construct. I’m sure someone with existing Atlas development knowledge could do this rather easily, but we’re willing to put in the effort if necessary.
My REAL question is this – what is the right way to get these changes integrated into the core product? I would be very sad to do a lot of work to integrate breed/species only to have Atlas 2.x come out an completely invalidate that work.
Is it possible to come to agreement on
a) how the data is stored
b) how the data is added to a cohort definition
Thanks All!
Chris - thanks for laying out the issues and I would support Option 2 as the most practical and viable. As with efforts to address adjacent clinical populations - I look forward to hearing what the Atlas/Tools team suggests as a long term approach to support in a sense “plug-ins” to enable utilizing adjacent clinical data to the core human CDM. I wonder if we can conceptually adopt what mobile device applications developers (ex Java J2ME) utilize - different profiles that help describe the target - in this case the core CDM and additional support tables. That is a Human CDM Profile and a Veterinary CDM Profile.
I’m not sure we need both Species and Breed columns.
Species is an ancestor of Breed concepts, please look here:
select * from concept_ancestor
join concept on concept_id = descendant_concept_id
where ancestor_concept_id = 40491509 -- Canis lupus familiaris
order by min_levels_of_separation
;
select * from concept_ancestor
join concept on concept_id = descendant_concept_id
where ancestor_concept_id = 40488452 -- Felis catus
order by min_levels_of_separation
the output (descendant_concept_id) has all dog and cat breeds, while the ancestor concept stands for Species.
You are bringing up an important point: How can we tweak ATLAS to a special use case that the community overall doesn’t care about, without cluttering it completely. This is a question for the folks who are thinking about the architecture and approach to Open Source development.
Tagging @gregk, @Chris_Knoll, @Frank, @anthonysena and @Ajit_Londhe. Can you guys talk to Chris?
To your questions, @Chris_Brandt:
- I’d also think Option 2. The “human” community shouldn’t be confused with breeds, and the “animal” community can tolerate the semantic abberation of calling an animal a person. After all, we are all animals, too.

- Cohorts are stored with subject_id, cohort_id (containing the definition), start_date and end_date. The subject_id contains the person_id. So, you don’t have a problem here.
Hi @waydes, @Chris_Brandt, et al.,
I’m starting to explore the possibility of using the CDM and ATLAS with animal health data and am wondering if there is additional progress or experience that you can share since this discussion? Did any of you try out option 2 and have success?