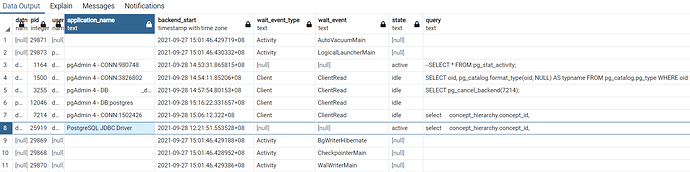
We are stuck in the drug treemap generation at 0%. I left the script running over the weekend and after 68 hours it is still executing the following SELECT statement:
select concept_hierarchy.concept_id,
COALESCE(concept_hierarchy.atc1_concept_name,'NA') || '||' ||
COALESCE(concept_hierarchy.atc3_concept_name,'NA') || '||' ||
COALESCE(concept_hierarchy.atc5_concept_name,'NA') || '||' ||
COALESCE(concept_hierarchy.rxnorm_ingredient_concept_name,'NA') || '||' ||
concept_hierarchy.rxnorm_concept_name concept_path,
ar1.count_value as num_persons,
ROUND(CAST(1.0*ar1.count_value / denom.count_value AS NUMERIC),5) as percent_persons,
ROUND(CAST(1.0*ar2.count_value / ar1.count_value AS NUMERIC),5) as records_per_person
from (select * from ACHILLES_results where analysis_id = 700) ar1
inner join
(select * from ACHILLES_results where analysis_id = 701) ar2
on ar1.stratum_1 = ar2.stratum_1
inner join
(
select rxnorm.concept_id,
rxnorm.concept_name as rxnorm_concept_name,
rxnorm.rxnorm_ingredient_concept_name,
atc5_to_atc3.atc5_concept_name,
atc3_to_atc1.atc3_concept_name,
atc1.concept_name as atc1_concept_name
from
(
select c1.concept_id,
c1.concept_name,
c2.concept_id as rxnorm_ingredient_concept_id,
c2.concept_name as RxNorm_ingredient_concept_name
from ohdsiv5.concept c1
inner join ohdsiv5.concept_ancestor ca1
on c1.concept_id = ca1.descendant_concept_id
and c1.vocabulary_id = 'RxNorm'
inner join ohdsiv5.concept c2
on ca1.ancestor_concept_id = c2.concept_id
and c2.vocabulary_id = 'RxNorm'
and c2.concept_class_id = 'Ingredient'
) rxnorm
left join
(select c1.concept_id as rxnorm_ingredient_concept_id, max(c2.concept_id) as atc5_concept_id
from
ohdsiv5.concept c1
inner join
ohdsiv5.concept_ancestor ca1
on c1.concept_id = ca1.descendant_concept_id
and c1.vocabulary_id = 'RxNorm'
and c1.concept_class_id = 'Ingredient'
inner join
ohdsiv5.concept c2
on ca1.ancestor_concept_id = c2.concept_id
and c2.vocabulary_id = 'ATC'
and c2.concept_class_id = 'ATC 4th'
group by c1.concept_id
) rxnorm_to_atc5
on rxnorm.rxnorm_ingredient_concept_id = rxnorm_to_atc5.rxnorm_ingredient_concept_id
left join
(select c1.concept_id as atc5_concept_id, c1.concept_name as atc5_concept_name, max(c2.concept_id) as atc3_concept_id
from
ohdsiv5.concept c1
inner join
ohdsiv5.concept_ancestor ca1
on c1.concept_id = ca1.descendant_concept_id
and c1.vocabulary_id = 'ATC'
and c1.concept_class_id = 'ATC 4th'
inner join
ohdsiv5.concept c2
on ca1.ancestor_concept_id = c2.concept_id
and c2.vocabulary_id = 'ATC'
and c2.concept_class_id = 'ATC 2nd'
group by c1.concept_id, c1.concept_name
) atc5_to_atc3
on rxnorm_to_atc5.atc5_concept_id = atc5_to_atc3.atc5_concept_id
left join
(select c1.concept_id as atc3_concept_id, c1.concept_name as atc3_concept_name, max(c2.concept_id) as atc1_concept_id
from
ohdsiv5.concept c1
inner join
ohdsiv5.concept_ancestor ca1
on c1.concept_id = ca1.descendant_concept_id
and c1.vocabulary_id = 'ATC'
and c1.concept_class_id = 'ATC 2nd'
inner join
ohdsiv5.concept c2
on ca1.ancestor_concept_id = c2.concept_id
and c2.vocabulary_id = 'ATC'
and c2.concept_class_id = 'ATC 1st'
group by c1.concept_id, c1.concept_name
) atc3_to_atc1
on atc5_to_atc3.atc3_concept_id = atc3_to_atc1.atc3_concept_id
left join ohdsiv5.concept atc1
on atc3_to_atc1.atc1_concept_id = atc1.concept_id
) concept_hierarchy
on ar1.stratum_1 = CAST(concept_hierarchy.concept_id AS VARCHAR)
,
(select count_value from ACHILLES_results where analysis_id = 1) denom
order by ar1.count_value desc
Indexes are enabled and database is tuned for performance, anybody having the same issue?